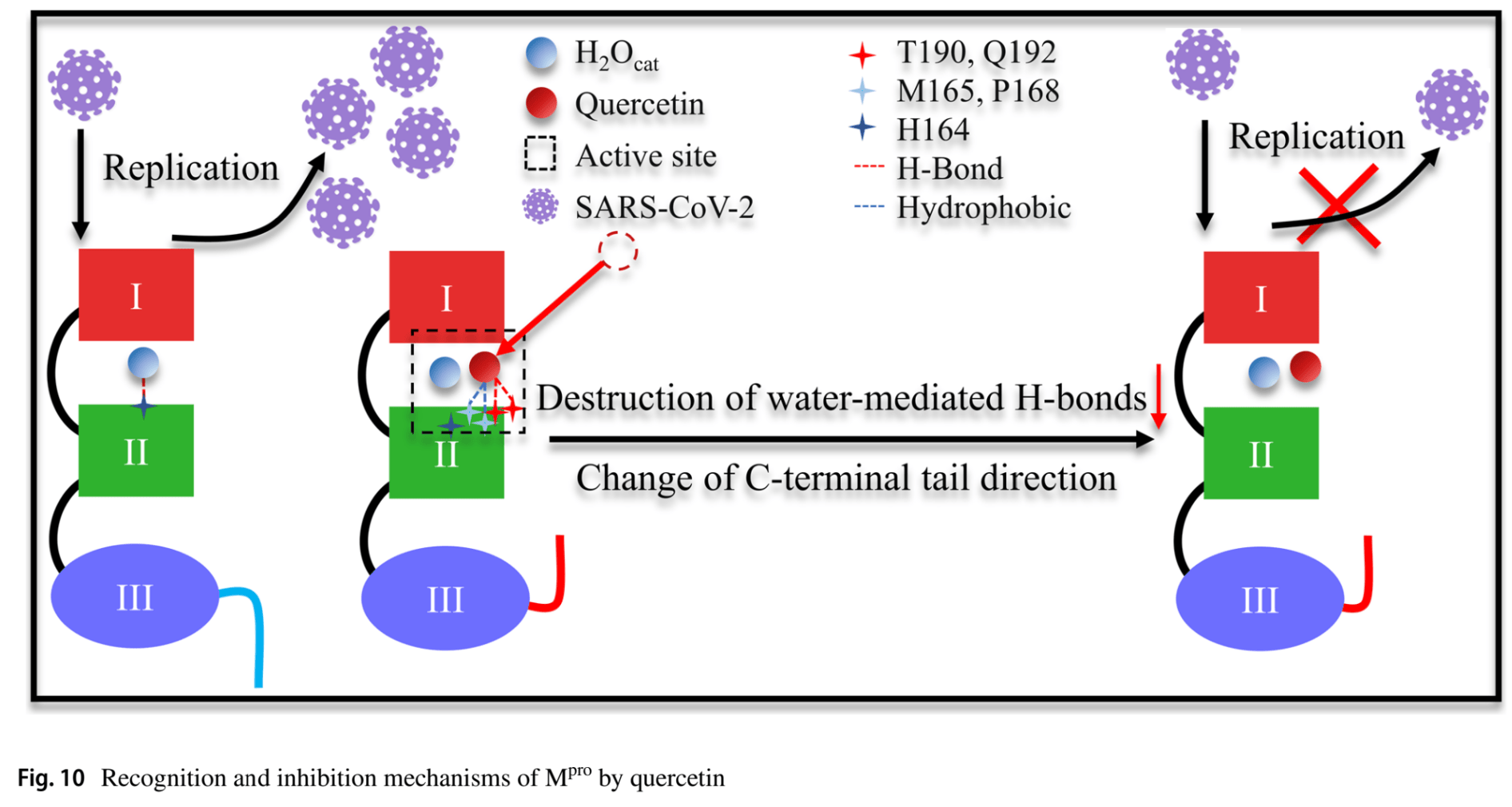
Feasibility of the inhibitor development for SARS-CoV-2: a systematic approach for drug design
et al., Journal of Molecular Modeling, doi:10.1007/s00894-025-06541-2, Nov 2025
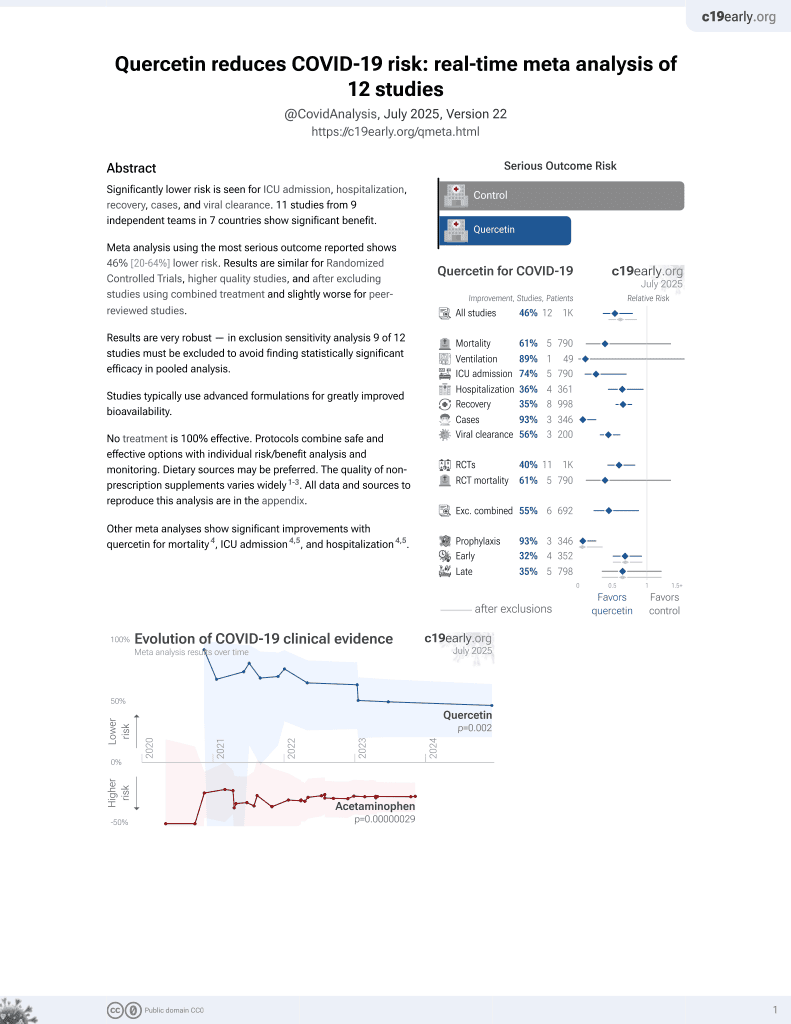
Quercetin for COVID-19
36th treatment shown to reduce risk in
January 2022, now with p = 0.0018 from 9 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico analysis identifying quercetin as a potent Mpro inhibitor through multi-stage virtual screening.
91 preclinical studies support the efficacy of quercetin for COVID-19:
In silico studies predict inhibition of SARS-CoV-2, or minimization of side effects, with quercetin or metabolites via binding to the spikeA,11,12,18,19,32,34,35,37,40,48,49,51,52,75 (and specifically the receptor binding domainB,8), MproC,7,8,11,12,16,18,20,22,24,26,28,30,33,34,37,40,44,46-48,52-55,72 , RNA-dependent RNA polymeraseD,8,10-12,18,42 , PLproE,12,47,55 , ACE2F,27,32,33,37,38,47,51 , TMPRSS2G,32, nucleocapsidH,12, helicaseI,12,39,44 , endoribonucleaseJ,49, NSP16/10K,15, cathepsin LL,36, Wnt-3M,32, FZDN,32, LRP6O,32, ezrinP,50, ADRPQ,48, NRP1R,51, EP300S,25, PTGS2T,33, HSP90AA1U,25,33 , matrix metalloproteinase 9V,41, IL-6W,31,45 , IL-10X,31, VEGFAY,45, and RELAZ,45 proteins, and inhibition of spike-ACE2 interactionAA,9.
In vitro studies demonstrate inhibition of the MproC,24,58,63,71 protein, and inhibition of spike-ACE2 interactionAA,59.
In vitro studies demonstrate efficacy in Calu-3AB,62, A549AC,31, HEK293-ACE2+AD,70, Huh-7AE,35, Caco-2AF,61, Vero E6AG,29,52,61 , mTECAH,64, RAW264.7AI,64, and HLMECAJ,9 cells.
Animal studies demonstrate efficacy in K18-hACE2 miceAK,67, db/db miceAL,64,74 , BALB/c miceAM,73, and rats29.
Quercetin reduced proinflammatory cytokines and protected lung and kidney tissue against LPS-induced damage in mice73, inhibits LPS-induced cytokine storm by modulating key inflammatory and antioxidant pathways in macrophages14, may block ACE2-spike interaction and NLRP3 inflammasome, limiting viral entry and inflammation5, upregulates the SIRT1/AMPK axis to inhibit oxidative injury and accelerate viral clearance76, inhibits SARS-CoV-2 ORF3a ion channel activity, which contributes to viral pathogenicity and cytotoxicity66, may alleviate COVID-19 ARDS via inhibition of EGFR and JAK2 inflammatory targets1, may destabilize the Spike protein, IL-6R, and integrins via conserved residues, blocking viral entry, hyperinflammation, and platelet aggregation77, and may reduce COVID-19 neuroinflammation and cognitive dysfunction through anti-inflammatory mechanisms and neuroprotective effects78.
1.
Gupta et al., Harnessing phytoconstituents to treat COVID-19 triggered acute respiratory distress syndrome: Insights from network pharmacology, and molecular modeling, Phytochemistry Letters, doi:10.1016/j.phytol.2025.104105.
2.
Sun et al., Feasibility of the inhibitor development for SARS-CoV-2: a systematic approach for drug design, Journal of Molecular Modeling, doi:10.1007/s00894-025-06541-2.
3.
Torabfam et al., Improving quercetin solubility via structural modification enhances dual-target coronavirus entry: an integrated in-vitro and in-silico study, Scientific Reports, doi:10.1038/s41598-025-27374-2.
4.
Abdelhameed et al., Phytochemical and antiviral investigation of Cynanchum acutum L. extract and derived semi-synthetic analogs targeting SARS-CoV-2 main protease, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-025-00907-2.
5.
Manikyam et al., INP-Guided Network Pharmacology Discloses Multi-Target Therapeutic Strategy Against Cytokine and IgE Storms in the SARS-CoV-2 NB.1.8.1 Variant, Research Square, doi:10.21203/rs.3.rs-6819274/v1.
6.
Makoana et al., Integration of metabolomics and chemometrics with in-silico and in-vitro approaches to unravel SARS-Cov-2 inhibitors from South African plants, PLOS ONE, doi:10.1371/journal.pone.0320415.
7.
Bano et al., Biochemical Screening of Phytochemicals and Identification of Scopoletin as a Potential Inhibitor of SARS-CoV-2 Mpro, Revealing Its Biophysical Impact on Structural Stability, Viruses, doi:10.3390/v17030402.
8.
Rajamanickam et al., Exploring the Potential of Siddha Formulation MilagaiKudineer-Derived Phytotherapeutics Against SARS-CoV-2: An In-Silico Investigation for Antiviral Intervention, Journal of Pharmacy and Pharmacology Research, doi:10.26502/fjppr.0105.
9.
Moharram et al., Secondary metabolites of Alternaria alternate appraisal of their SARS-CoV-2 inhibitory and anti-inflammatory potentials, PLOS ONE, doi:10.1371/journal.pone.0313616.
10.
Metwaly et al., Integrated study of Quercetin as a potent SARS-CoV-2 RdRp inhibitor: Binding interactions, MD simulations, and In vitro assays, PLOS ONE, doi:10.1371/journal.pone.0312866.
11.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
12.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
13.
Pan et al., Decoding the mechanism of Qingjie formula in the prevention of COVID-19 based on network pharmacology and molecular docking, Heliyon, doi:10.1016/j.heliyon.2024.e39167.
14.
Xu et al., Quercetin inhibited LPS-induced cytokine storm by interacting with the AKT1-FoxO1 and Keap1-Nrf2 signaling pathway in macrophages, Scientific Reports, doi:10.1038/s41598-024-71569-y.
15.
Tamil Selvan et al., Computational Investigations to Identify Potent Natural Flavonoid Inhibitors of the Nonstructural Protein (NSP) 16/10 Complex Against Coronavirus, Cureus, doi:10.7759/cureus.68098.
16.
Sunita et al., Characterization of Phytochemical Inhibitors of the COVID-19 Primary Protease Using Molecular Modelling Approach, Asian Journal of Microbiology and Biotechnology, doi:10.56557/ajmab/2024/v9i28800.
17.
Wu et al., Biomarkers Prediction and Immune Landscape in Covid-19 and “Brain Fog”, Elsevier BV, doi:10.2139/ssrn.4897774.
18.
Raman et al., Phytoconstituents of Citrus limon (Lemon) as Potential Inhibitors Against Multi Targets of SARS‐CoV‐2 by Use of Molecular Modelling and In Vitro Determination Approaches, ChemistryOpen, doi:10.1002/open.202300198.
19.
Asad et al., Exploring the antiviral activity of Adhatoda beddomei bioactive compounds in interaction with coronavirus spike protein, Archives of Medical Reports, 1:1, archmedrep.com/index.php/amr/article/view/3.
20.
Irfan et al., Phytoconstituents of Artemisia Annua as potential inhibitors of SARS CoV2 main protease: an in silico study, BMC Infectious Diseases, doi:10.1186/s12879-024-09387-w.
21.
Yuan et al., Network pharmacology and molecular docking reveal the mechanisms of action of Panax notoginseng against post-COVID-19 thromboembolism, Review of Clinical Pharmacology and Pharmacokinetics - International Edition, doi:10.61873/DTFA3974.
22.
Nalban et al., Targeting COVID-19 (SARS-CoV-2) main protease through phytochemicals of Albizia lebbeck: molecular docking, molecular dynamics simulation, MM–PBSA free energy calculations, and DFT analysis, Journal of Proteins and Proteomics, doi:10.1007/s42485-024-00136-w.
23.
Zhou et al., Bioinformatics and system biology approaches to determine the connection of SARS-CoV-2 infection and intrahepatic cholangiocarcinoma, PLOS ONE, doi:10.1371/journal.pone.0300441.
24.
Waqas et al., Discovery of Novel Natural Inhibitors Against SARS-CoV-2 Main Protease: A Rational Approach to Antiviral Therapeutics, Current Medicinal Chemistry, doi:10.2174/0109298673292839240329081008.
25.
Hasanah et al., Decoding the therapeutic potential of empon-empon: a bioinformatics expedition unraveling mechanisms against COVID-19 and atherosclerosis, International Journal of Applied Pharmaceutics, doi:10.22159/ijap.2024v16i2.50128.
26.
Shaik et al., Computational identification of selected bioactive compounds from Cedrus deodara as inhibitors against SARS-CoV-2 main protease: a pharmacoinformatics study, Indian Drugs, doi:10.53879/id.61.02.13859.
27.
Wang et al., Investigating the Mechanism of Qu Du Qiang Fei 1 Hao Fang Formula against Coronavirus Disease 2019 Based on Network Pharmacology Method, World Journal of Traditional Chinese Medicine, doi:10.4103/2311-8571.395061.
28.
Singh et al., Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach, Research Square, doi:10.21203/rs.3.rs-3888947/v1.
29.
El-Megharbel et al., Chemical and spectroscopic characterization of (Artemisinin/Quercetin/ Zinc) novel mixed ligand complex with assessment of its potent high antiviral activity against SARS-CoV-2 and antioxidant capacity against toxicity induced by acrylamide in male rats, PeerJ, doi:10.7717/peerj.15638.
30.
Akinwumi et al., Evaluation of therapeutic potentials of some bioactive compounds in selected African plants targeting main protease (Mpro) in SARS-CoV-2: a molecular docking study, Egyptian Journal of Medical Human Genetics, doi:10.1186/s43042-023-00456-4.
31.
Yang et al., Active ingredient and mechanistic analysis of traditional Chinese medicine formulas for the prevention and treatment of COVID-19: Insights from bioinformatics and in vitro experiments, Medicine, doi:10.1097/MD.0000000000036238.
32.
Chandran et al., Molecular docking analysis of quercetin with known CoVid-19 targets, Bioinformation, doi:10.6026/973206300191081.
33.
Qin et al., Exploring the bioactive compounds of Feiduqing formula for the prevention and management of COVID-19 through network pharmacology and molecular docking, Medical Data Mining, doi:10.53388/MDM202407003.
34.
Moschovou et al., Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms242115894.
35.
Pan (B) et al., Quercetin: A promising drug candidate against the potential SARS-CoV-2-Spike mutants with high viral infectivity, Computational and Structural Biotechnology Journal, doi:10.1016/j.csbj.2023.10.029.
36.
Ahmed et al., Evaluation of the Effect of Zinc, Quercetin, Bromelain and Vitamin C on COVID-19 Patients, International Journal of Diabetes Management, doi:10.61797/ijdm.v2i2.259.
37.
Thapa et al., In-silico Approach for Predicting the Inhibitory Effect of Home Remedies on Severe Acute Respiratory Syndrome Coronavirus-2, Makara Journal of Science, doi:10.7454/mss.v27i3.1609.
38.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
39.
Singh (B) et al., Flavonoids as Potent Inhibitor of SARS-CoV-2 Nsp13 Helicase: Grid Based Docking Approach, Middle East Research Journal of Pharmaceutical Sciences, doi:10.36348/merjps.2023.v03i04.001.
40.
Mandal et al., In silico anti-viral assessment of phytoconstituents in a traditional (Siddha Medicine) polyherbal formulation – Targeting Mpro and pan-coronavirus post-fusion Spike protein, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2023.07.004.
41.
Sai Ramesh et al., Computational analysis of the phytocompounds of Mimusops elengi against spike protein of SARS CoV2 – An Insilico model, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2023.125553.
42.
Corbo et al., Inhibitory potential of phytochemicals on five SARS-CoV-2 proteins: in silico evaluation of endemic plants of Bosnia and Herzegovina, Biotechnology & Biotechnological Equipment, doi:10.1080/13102818.2023.2222196.
43.
Azmi et al., Utilization of quercetin flavonoid compounds in onion (Allium cepa L.) as an inhibitor of SARS-CoV-2 spike protein against ACE2 receptors, 11th International Seminar on New Paradigm and Innovation on Natural Sciences and its Application, doi:10.1063/5.0140285.
44.
Alanzi et al., Structure-based virtual identification of natural inhibitors of SARS-CoV-2 and its Delta and Omicron variant proteins, Future Virology, doi:10.2217/fvl-2022-0184.
45.
Yang (B) et al., In silico evidence implicating novel mechanisms of Prunella vulgaris L. as a potential botanical drug against COVID-19-associated acute kidney injury, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1188086.
46.
Wang (B) et al., Computational Analysis of Lianhua Qingwen as an Adjuvant Treatment in Patients with COVID-19, Society of Toxicology Conference, 2023, www.researchgate.net/publication/370491709_Y_Wang_A_E_Tan_O_Chew_A_Hsueh_and_D_E_Johnson_2023_Computational_Analysis_of_Lianhua_Qingwen_as_an_Adjuvant_Treatment_in_Patients_with_COVID-19_Toxicologist_1921_507.
47.
Ibeh et al., Computational studies of potential antiviral compounds from some selected Nigerian medicinal plants against SARS-CoV-2 proteins, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2023.101230.
48.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
49.
Alavi et al., Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study, Biomedicines, doi:10.3390/biomedicines10123074.
50.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
51.
Şimşek et al., In silico identification of SARS-CoV-2 cell entry inhibitors from selected natural antivirals, Journal of Molecular Graphics and Modelling, doi:10.1016/j.jmgm.2021.108038.
52.
Kandeil et al., Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2, Pathogens, doi:10.3390/pathogens10060758.
53.
Rehman et al., Natural Compounds as Inhibitors of SARS-CoV-2 Main Protease (3CLpro): A Molecular Docking and Simulation Approach to Combat COVID-19, Current Pharmaceutical Design, doi:10.2174/1381612826999201116195851.
54.
Sekiou et al., In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2, ChemRxiv, doi:10.26434/chemrxiv.12181404.v1.
55.
Zhang et al., In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus, Journal of Integrative Medicine, doi:10.1016/j.joim.2020.02.005.
56.
Sisti et al., Evaluation of respiratory virus transmissibility and resilience from fomites: the case of 11 SARS-CoV-2 clinical isolates, Applied and Environmental Microbiology, doi:10.1128/aem.00774-25.
57.
Spinelli et al., Amphibian‐Derived Peptides as Natural Inhibitors of SARS‐CoV‐2 Main Protease (Mpro): A Combined In Vitro and In Silico Approach, Chemistry & Biodiversity, doi:10.1002/cbdv.202403202.
58.
Aguilera-Rodriguez et al., Inhibition of SARS-CoV-2 3CLpro by chemically modified tyrosinase from Agaricus bisporus, RSC Medicinal Chemistry, doi:10.1039/D4MD00289J.
59.
Emam et al., Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors, AMB Express, doi:10.1186/s13568-024-01739-8.
60.
Fang et al., Development of nanoparticles incorporated with quercetin and ACE2-membrane as a novel therapy for COVID-19, Journal of Nanobiotechnology, doi:10.1186/s12951-024-02435-2.
61.
Roy et al., Quercetin inhibits SARS-CoV-2 infection and prevents syncytium formation by cells co-expressing the viral spike protein and human ACE2, Virology Journal, doi:10.1186/s12985-024-02299-w.
62.
DiGuilio et al., Quercetin improves and protects Calu-3 airway epithelial barrier function, Frontiers in Cell and Developmental Biology, doi:10.3389/fcell.2023.1271201.
63.
Zhang (B) et al., Discovery of the covalent SARS‐CoV‐2 Mpro inhibitors from antiviral herbs via integrating target‐based high‐throughput screening and chemoproteomic approaches, Journal of Medical Virology, doi:10.1002/jmv.29208.
64.
Wu (B) et al., SARS-CoV-2 N protein induced acute kidney injury in diabetic db/db mice is associated with a Mincle-dependent M1 macrophage activation, Frontiers in Immunology, doi:10.3389/fimmu.2023.1264447.
65.
Xu (B) et al., Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19, Proceedings of the National Academy of Sciences, doi:10.1073/pnas.2301775120.
66.
Fam et al., Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Scientific Reports, doi:10.1038/s41598-023-31764-9.
67.
Aguado et al., Senolytic therapy alleviates physiological human brain aging and COVID-19 neuropathology, bioRxiv, doi:10.1101/2023.01.17.524329.
68.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
69.
Munafò et al., Quercetin and Luteolin Are Single-digit Micromolar Inhibitors of the SARS-CoV-2 RNA-dependent RNA Polymerase, Research Square, doi:10.21203/rs.3.rs-1149846/v1.
70.
Singh (C) et al., The spike protein of SARS-CoV-2 virus induces heme oxygenase-1: Pathophysiologic implications, Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, doi:10.1016/j.bbadis.2021.166322.
71.
Bahun et al., Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols, Food Chemistry, doi:10.1016/j.foodchem.2021.131594.
72.
Abian et al., Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.07.235.
73.
Shaker et al., Anti-cytokine Storm Activity of Fraxin, Quercetin, and their Combination on Lipopolysaccharide-Induced Cytokine Storm in Mice: Implications in COVID-19, Iranian Journal of Medical Sciences, doi:10.30476/ijms.2023.98947.3102.
74.
Wu (C) et al., Treatment with Quercetin inhibits SARS-CoV-2 N protein-induced acute kidney injury by blocking Smad3-dependent G1 cell cycle arrest, Molecular Therapy, doi:10.1016/j.ymthe.2022.12.002.
75.
Azmi (B) et al., The role of vitamin D receptor and IL‐6 in COVID‐19, Molecular Genetics & Genomic Medicine, doi:10.1002/mgg3.2172.
76.
Shokri-Afra et al., Targeting SIRT1: A Potential Strategy for Combating Severe COVID‐19, BioMed Research International, doi:10.1155/bmri/9507417.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
g.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
h.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
i.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
j.
The endoribonuclease, also known as NendoU or nsp15, cleaves specific sequences in viral RNA which may help the virus evade detection by the host immune system. Inhibition may hinder the virus's ability to mask itself from the immune system, facilitating a stronger immune response.
k.
The NSP16/10 complex consists of non-structural proteins 16 and 10, forming a 2'-O-methyltransferase that modifies the viral RNA cap structure. This modification helps the virus evade host immune detection by mimicking host mRNA, making NSP16/10 a promising antiviral target.
l.
Cathepsin L is a host lysosomal cysteine protease that can prime the spike protein through an alternative pathway when TMPRSS2 is unavailable. Dual targeting of cathepsin L and TMPRSS2 may maximize disruption of alternative pathways for virus entry.
m.
Wingless-related integration site (Wnt) ligand 3 is a host signaling molecule that activates the Wnt signaling pathway, which is important in development, cell growth, and tissue repair. Some studies suggest that SARS-CoV-2 infection may interfere with the Wnt signaling pathway, and that Wnt3a is involved in SARS-CoV-2 entry.
n.
The frizzled (FZD) receptor is a host transmembrane receptor that binds Wnt ligands, initiating the Wnt signaling cascade. FZD serves as a co-receptor, along with ACE2, in some proposed mechanisms of SARS-CoV-2 infection. The virus may take advantage of this pathway as an alternative entry route.
o.
Low-density lipoprotein receptor-related protein 6 is a cell surface co-receptor essential for Wnt signaling. LRP6 acts in tandem with FZD for signal transduction and has been discussed as a potential co-receptor for SARS-CoV-2 entry.
p.
The ezrin protein links the cell membrane to the cytoskeleton (the cell's internal support structure) and plays a role in cell shape, movement, adhesion, and signaling. Drugs that occupy the same spot on ezrin where the viral spike protein would bind may hindering viral attachment, and drug binding could further stabilize ezrin, strengthening its potential natural capacity to impede viral fusion and entry.
q.
The Adipocyte Differentiation-Related Protein (ADRP, also known as Perilipin 2 or PLIN2) is a lipid droplet protein regulating the storage and breakdown of fats in cells. SARS-CoV-2 may hijack the lipid handling machinery of host cells and ADRP may play a role in this process. Disrupting ADRP's interaction with the virus may hinder the virus's ability to use lipids for replication and assembly.
r.
Neuropilin-1 (NRP1) is a cell surface receptor with roles in blood vessel development, nerve cell guidance, and immune responses. NRP1 may function as a co-receptor for SARS-CoV-2, facilitating viral entry into cells. Blocking NRP1 may disrupt an alternative route of viral entry.
s.
EP300 (E1A Binding Protein P300) is a transcriptional coactivator involved in several cellular processes, including growth, differentiation, and apoptosis, through its acetyltransferase activity that modifies histones and non-histone proteins. EP300 facilitates viral entry into cells and upregulates inflammatory cytokine production.
t.
Prostaglandin G/H synthase 2 (PTGS2, also known as COX-2) is an enzyme crucial for the production of inflammatory molecules called prostaglandins. PTGS2 plays a role in the inflammatory response that can become severe in COVID-19 and inhibitors (like some NSAIDs) may have benefits in dampening harmful inflammation, but note that prostaglandins have diverse physiological functions.
u.
Heat Shock Protein 90 Alpha Family Class A Member 1 (HSP90AA1) is a chaperone protein that helps other proteins fold correctly and maintains their stability. HSP90AA1 plays roles in cell signaling, survival, and immune responses. HSP90AA1 may interact with numerous viral proteins, but note that it has diverse physiological functions.
v.
Matrix metalloproteinase 9 (MMP9), also called gelatinase B, is a zinc-dependent enzyme that breaks down collagen and other components of the extracellular matrix. MMP9 levels increase in severe COVID-19. Overactive MMP9 can damage lung tissue and worsen inflammation. Inhibition of MMP9 may prevent excessive tissue damage and help regulate the inflammatory response.
w.
The interleukin-6 (IL-6) pro-inflammatory cytokine (signaling molecule) has a complex role in the immune response and may trigger and perpetuate inflammation. Elevated IL-6 levels are associated with severe COVID-19 cases and cytokine storm. Anti-IL-6 therapies may be beneficial in reducing excessive inflammation in severe COVID-19 cases.
x.
The interleukin-10 (IL-10) anti-inflammatory cytokine helps regulate and dampen immune responses, preventing excessive inflammation. IL-10 levels can also be elevated in severe COVID-19. IL-10 could either help control harmful inflammation or potentially contribute to immune suppression.
y.
Vascular Endothelial Growth Factor A (VEGFA) promotes the growth of new blood vessels (angiogenesis) and has roles in inflammation and immune responses. VEGFA may contribute to blood vessel leakiness and excessive inflammation associated with severe COVID-19.
z.
RELA is a transcription factor subunit of NF-kB and is a key regulator of inflammation, driving pro-inflammatory gene expression. SARS-CoV-2 may hijack and modulate NF-kB pathways.
aa.
The interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor is a primary method of viral entry, inhibiting this interaction can prevent the virus from attaching to and entering host cells, halting infection at an early stage.
ab.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
ac.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
ad.
HEK293-ACE2+ is a human embryonic kidney cell line engineered for high ACE2 expression and SARS-CoV-2 susceptibility.
ae.
Huh-7 cells were derived from a liver tumor (hepatoma).
af.
Caco-2 cells come from a colorectal adenocarcinoma (cancer). They are valued for their ability to form a polarized cell layer with properties similar to the intestinal lining.
ag.
Vero E6 is an African green monkey kidney cell line with low/no ACE2 expression and high SARS-CoV-2 susceptibility. The cell line is easy to maintain and supports robust viral replication, however the monkey origin may not accurately represent human responses.
ah.
mTEC is a mouse tubular epithelial cell line.
ai.
RAW264.7 is a mouse macrophage cell line.
aj.
HLMEC (Human Lung Microvascular Endothelial Cells) are primary endothelial cells derived from the lung microvasculature. They are used to study endothelial function, inflammation, and viral interactions, particularly in the context of lung infections such as SARS-CoV-2. HLMEC express ACE2 and are susceptible to SARS-CoV-2 infection, making them a relevant model for studying viral entry and endothelial responses in the lung.
ak.
A mouse model expressing the human ACE2 receptor under the control of the K18 promoter.
al.
A mouse model of obesity and severe insulin resistance leading to type 2 diabetes due to a mutation in the leptin receptor gene that impairs satiety signaling.
am.
A mouse model commonly used in infectious disease and cancer research due to higher immune response and susceptibility to infection.
Sun et al., 28 Nov 2025, peer-reviewed, 10 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
DOI record:
{
"DOI": "10.1007/s00894-025-06541-2",
"ISSN": [
"1610-2940",
"0948-5023"
],
"URL": "http://dx.doi.org/10.1007/s00894-025-06541-2",
"alternative-id": [
"6541"
],
"article-number": "352",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "2 September 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2 October 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "28 November 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Date",
"name": "change_date",
"order": 5,
"value": "19 December 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Type",
"name": "change_type",
"order": 6,
"value": "Update"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Details",
"name": "change_details",
"order": 7,
"value": "The original online version of this article was revised to correct the affiliations of author."
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Ethics approval and consent to participate",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "Not applicable."
},
{
"group": {
"label": "Consent for publication",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 3,
"value": "Not applicable."
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 4,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Sun",
"given": "Guangzhou",
"sequence": "first"
},
{
"affiliation": [],
"family": "Shi",
"given": "Quanshan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Song",
"given": "Yuting",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cheng",
"given": "Dazhi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jiang",
"given": "Yu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hu",
"given": "Dongling",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yue",
"given": "Xinru",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yu",
"given": "Wentong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shi",
"given": "Xiaodong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hu",
"given": "Jianping",
"sequence": "additional"
}
],
"container-title": "Journal of Molecular Modeling",
"container-title-short": "J Mol Model",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
11,
28
]
],
"date-time": "2025-11-28T06:29:25Z",
"timestamp": 1764311365000
},
"deposited": {
"date-parts": [
[
2026,
1,
5
]
],
"date-time": "2026-01-05T18:34:45Z",
"timestamp": 1767638085000
},
"indexed": {
"date-parts": [
[
2026,
1,
5
]
],
"date-time": "2026-01-05T20:51:06Z",
"timestamp": 1767646266961,
"version": "3.48.0"
},
"is-referenced-by-count": 0,
"issue": "12",
"issued": {
"date-parts": [
[
2025,
11,
28
]
]
},
"journal-issue": {
"issue": "12",
"published-print": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.springernature.com/gp/researchers/text-and-data-mining",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
11,
28
]
],
"date-time": "2025-11-28T00:00:00Z",
"timestamp": 1764288000000
}
},
{
"URL": "https://www.springernature.com/gp/researchers/text-and-data-mining",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
11,
28
]
],
"date-time": "2025-11-28T00:00:00Z",
"timestamp": 1764288000000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s00894-025-06541-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s00894-025-06541-2",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s00894-025-06541-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1007",
"published": {
"date-parts": [
[
2025,
11,
28
]
]
},
"published-online": {
"date-parts": [
[
2025,
11,
28
]
]
},
"published-print": {
"date-parts": [
[
2025,
12
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1038/s41586-020-2202-3",
"author": "F Wu",
"doi-asserted-by": "publisher",
"first-page": "265",
"issue": "7798",
"journal-title": "Nature",
"key": "6541_CR1",
"unstructured": "Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY et al (2020) A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269. https://doi.org/10.1038/s41586-020-2202-3",
"volume": "579",
"year": "2020"
},
{
"key": "6541_CR2",
"unstructured": "World Health Organization (2020) Dépistage en laboratoire des cas suspects d’infection humaine par le nouveau coronavirus 2019 (2019-nCoV): lignes directrices provisoires, 17 janvier 2020[M]. World Health Organization. https://covid19.who.int/ . Accessed 15 Jan 2022"
},
{
"DOI": "10.1016/S2213-2600(20)30370-2",
"author": "G Grasselli",
"doi-asserted-by": "publisher",
"first-page": "1201",
"issue": "12",
"journal-title": "The lancet Resp med",
"key": "6541_CR3",
"unstructured": "Grasselli G, Tonetti T, Protti A, Langer T, Girardis M, Bellani G, Laggey J, Carrafiello PG, Carasana L, Rizzuto C et al (2020) Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. The lancet Resp med 8(12):1201–1208. https://doi.org/10.1016/S2213-2600(20)30370-2",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.2139/ssrn.3566298",
"doi-asserted-by": "crossref",
"key": "6541_CR4",
"unstructured": "Jebril1a NMT(2020) World Health Organization declared a pandemic public health menace: a systematic review of the coronavirus disease 2019 “COVID-19”. International Journal of Psychosocial Rehabilitation 24(9). https://www.psychosocial.com/article/PR290311/25748/"
},
{
"DOI": "10.1001/jama.2020.1585",
"author": "D Wang",
"doi-asserted-by": "publisher",
"first-page": "1061",
"issue": "11",
"journal-title": "JAMA",
"key": "6541_CR5",
"unstructured": "Wang D, Hu B, Hu C, Zhu FF, Liu X, Zhang J, Wang BB, Xiang H, Cheng ZS, Xiong Y et al (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan China. JAMA 323(11):1061–1069. https://doi.org/10.1001/jama.2020.1585",
"volume": "323",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2739-1",
"author": "Y Finkel",
"doi-asserted-by": "publisher",
"first-page": "125",
"issue": "7840",
"journal-title": "Nature",
"key": "6541_CR6",
"unstructured": "Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdoit H, Stein D, Israeli O et al (2021) The coding capacity of SARS-CoV-2. Nature 589(7840):125–130. https://doi.org/10.1038/s41586-020-2739-1",
"volume": "589",
"year": "2021"
},
{
"DOI": "10.1042/BCJ20200029",
"author": "B Krichel",
"doi-asserted-by": "publisher",
"first-page": "1009",
"issue": "5",
"journal-title": "Biochem J",
"key": "6541_CR7",
"unstructured": "Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C (2020) Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J 477(5):1009–1019. https://doi.org/10.1042/BCJ20200029",
"volume": "477",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"author": "P Zhou",
"doi-asserted-by": "publisher",
"first-page": "270",
"issue": "7798",
"journal-title": "Nature",
"key": "6541_CR8",
"unstructured": "Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2012-7",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1126/science.1085658",
"author": "K Anand",
"doi-asserted-by": "publisher",
"first-page": "1763",
"issue": "5626",
"journal-title": "Science",
"key": "6541_CR9",
"unstructured": "Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R (2003) Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300(5626):1763–1767. https://doi.org/10.1126/science.1085658",
"volume": "300",
"year": "2003"
},
{
"DOI": "10.1080/07391102.2021.1927845",
"author": "J Novak",
"doi-asserted-by": "publisher",
"first-page": "9347",
"issue": "19",
"journal-title": "J Biomol Struct Dyn",
"key": "6541_CR10",
"unstructured": "Novak J, Rimac H, Kandagalla S, Pathak P, Naumovich V, Grishina M, Potemkin V (2021) Proposition of a new allosteric binding site for potential SARS-CoV-2 3CL protease inhibitors by utilizing molecular dynamics simulations and ensemble docking. J Biomol Struct Dyn 40(19):9347–9360. https://doi.org/10.1080/07391102.2021.1927845",
"volume": "40",
"year": "2021"
},
{
"DOI": "10.1038/srep22677",
"author": "F Wang",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Sci Rep-UK",
"key": "6541_CR11",
"unstructured": "Wang F, Chen C, Tan W, Yang K, Yang H (2016) Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci Rep-UK 6(1):1–12. https://doi.org/10.1038/srep22677",
"volume": "6",
"year": "2016"
},
{
"DOI": "10.1038/d41587-020-00005-z",
"author": "J Hodgson",
"doi-asserted-by": "publisher",
"first-page": "523",
"issue": "5",
"journal-title": "Nat Biotechnol",
"key": "6541_CR12",
"unstructured": "Hodgson J (2020) The pandemic pipeline. Nat Biotechnol 38(5):523–532. https://doi.org/10.1038/d41587-020-00005-z",
"volume": "38",
"year": "2020"
},
{
"DOI": "10.1038/s41598-021-99165-4",
"doi-asserted-by": "crossref",
"key": "6541_CR13",
"unstructured": "Patel CN, Jani SP, Jaiswal DG, Kumar SP, Mangukia N, Parmar RM, ..., Pandya HA (2021) Identification of antiviral phytochemicals as a potential SARS-CoV-2 main protease (Mpro) inhibitor using docking and molecular dynamics simulations. Sci Rep 11(1):20295"
},
{
"DOI": "10.3389/fchem.2021.622898",
"author": "HM Mengist",
"doi-asserted-by": "publisher",
"journal-title": "Front Chem",
"key": "6541_CR14",
"unstructured": "Mengist HM, Dilnessa T, Jin T (2021) Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front Chem 9:622898. https://doi.org/10.3389/fchem.2021.622898",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1016/j.compbiomed.2022.106318",
"author": "CN Patel",
"doi-asserted-by": "publisher",
"journal-title": "Comput Biol Med",
"key": "6541_CR15",
"unstructured": "Patel CN, Jani SP, Kumar SP, Modi KM, Kumar Y (2022) Computational investigation of natural compounds as potential main protease (Mpro) inhibitors for SARS-CoV-2 virus. Comput Biol Med 151:106318. https://doi.org/10.1016/j.compbiomed.2022.106318",
"volume": "151",
"year": "2022"
},
{
"DOI": "10.1056/NEJMc2112981",
"author": "J Keehner",
"doi-asserted-by": "publisher",
"first-page": "1330",
"issue": "14",
"journal-title": "N Engl J Med",
"key": "6541_CR16",
"unstructured": "Keehner J, Horton LE, Binkin NJ, Laurent LC, Pride D, Longhurst CA, Abeles SR, Torriani FJ (2021) Resurgence of SARS-CoV-2 infection in a highly vaccinated health system workforce. N Engl J Med 385(14):1330–1332. https://doi.org/10.1056/NEJMc2112981",
"volume": "385",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"author": "Z Jin",
"doi-asserted-by": "publisher",
"first-page": "289",
"issue": "7811",
"journal-title": "Nature",
"key": "6541_CR17",
"unstructured": "Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C et al (2020) Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582(7811):289–293. https://doi.org/10.1038/s41586-020-2223-y",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1016/j.coviro.2021.04.006",
"author": "K Vandyck",
"doi-asserted-by": "publisher",
"first-page": "36",
"journal-title": "Curr Opin Virol",
"key": "6541_CR18",
"unstructured": "Vandyck K, Deval J (2021) Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr Opin Virol 49:36–40. https://doi.org/10.1016/j.coviro.2021.04.006",
"volume": "49",
"year": "2021"
},
{
"DOI": "10.1016/j.drup.2021.100794",
"author": "S Drożdżal",
"doi-asserted-by": "publisher",
"journal-title": "Drug Resist Update",
"key": "6541_CR19",
"unstructured": "Drożdżal S, Rosik J, Lechowicz K, Machaj F, Szostak B, Przybyciński J, Lorzadeh S, Kotfis K, Ghavami S, Los M (2021) An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Update 59:100794. https://doi.org/10.1016/j.drup.2021.100794",
"volume": "59",
"year": "2021"
},
{
"DOI": "10.1007/s13238-021-00883-2",
"author": "Y Zhao",
"doi-asserted-by": "publisher",
"first-page": "689",
"issue": "9",
"journal-title": "Protein Cell",
"key": "6541_CR20",
"unstructured": "Zhao Y, Fang C, Zhang Q, Zhang R, Zhao X, Duan Y, Wang H, Zhu Y, Feng L, Zhao J et al (2022) Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 13(9):689–693. https://doi.org/10.1007/s13238-021-00883-2",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1128/jvi.02013-21",
"author": "J Li",
"doi-asserted-by": "publisher",
"first-page": "e02013",
"issue": "8",
"journal-title": "J Virol",
"key": "6541_CR21",
"unstructured": "Li J, Lin C, Zhou X, Zhong F, Zeng P, Yang Y, Zhang Y, Yu B, Fan X, McCormick PJ et al (2022) Structural basis of the main proteases of coronavirus bound to drug candidate PF-07321332. J Virol 96(8):e02013–e02021. https://doi.org/10.1128/jvi.02013-21",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/j.compbiolchem.2022.107692",
"author": "YJ Alvarado",
"doi-asserted-by": "publisher",
"journal-title": "Comput Biol Chem",
"key": "6541_CR22",
"unstructured": "Alvarado YJ, Olivarez Y, Lossada C, Vera-Villalobos J, Paz JL, Vera E, Lorono M, Vivas A, Torres FJ, Jeffreys LN et al (2022) Interaction of the new inhibitor Paxlovid (PF-07321332) and ivermectin with the monomer of the main protease SARS-CoV-2: a volumetric study based on molecular dynamics Elastic Networks Classical Thermodynamics and Spt. Comput Biol Chem 99:107692. https://doi.org/10.1016/j.compbiolchem.2022.107692",
"volume": "99",
"year": "2022"
},
{
"DOI": "10.1016/j.jmgm.2021.108042",
"author": "M Macchiagodena",
"doi-asserted-by": "publisher",
"journal-title": "J Mol Graph Model",
"key": "6541_CR23",
"unstructured": "Macchiagodena M, Pagliai M, Procacci P (2022) Characterization of the non-covalent interaction between the PF-07321332 inhibitor and the SARS-CoV-2 main protease. J Mol Graph Model 110:108042. https://doi.org/10.1016/j.jmgm.2021.108042",
"volume": "110",
"year": "2022"
},
{
"DOI": "10.1016/j.bbrc.2020.08.116",
"author": "Y Jiang",
"doi-asserted-by": "publisher",
"first-page": "47",
"journal-title": "Biochem Biophys Res Commun",
"key": "6541_CR24",
"unstructured": "Jiang Y, Yin W, Xu HE (2021) RNA-dependent RNA polymerase: structure mechanism and drug discovery for COVID-19. Biochem Biophys Res Commun 538:47–53. https://doi.org/10.1016/j.bbrc.2020.08.116",
"volume": "538",
"year": "2021"
},
{
"DOI": "10.1111/ced.15281",
"author": "M Pupo Correia",
"doi-asserted-by": "publisher",
"first-page": "1738",
"issue": "9",
"journal-title": "Clin Exp Dermatol",
"key": "6541_CR25",
"unstructured": "Pupo Correia M, Fernandes S, Filipe P (2022) Cutaneous adverse reactions to the new oral antiviral drugs against SARS-CoV-2. Clin Exp Dermatol 47(9):1738–1740. https://doi.org/10.1111/ced.15281",
"volume": "47",
"year": "2022"
},
{
"DOI": "10.1021/acs.chemrev.1c00965",
"author": "K Gao",
"doi-asserted-by": "publisher",
"first-page": "11287",
"issue": "13",
"journal-title": "Chem Rev",
"key": "6541_CR26",
"unstructured": "Gao K, Wang R, Chen J, Cheng L, Frishcosy J, Huzumi Y, Qiu Y, Schluckbier T, Wei X, Wei GW (2022) Methodology-centered review of molecular modeling, simulation, and prediction of SARS-CoV-2. Chem Rev 122(13):11287–11368",
"volume": "122",
"year": "2022"
},
{
"DOI": "10.14218/JCTH.2021.00368",
"author": "R Zhang",
"doi-asserted-by": "publisher",
"first-page": "748",
"issue": "4",
"journal-title": "J Clin Transl Hepatol",
"key": "6541_CR27",
"unstructured": "Zhang R, Wang Q, Yang J (2022) Impact of liver functions by repurposed drugs for COVID-19 treatment. J Clin Transl Hepatol 10(4):748–756. https://doi.org/10.14218/JCTH.2021.00368",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.3390/audiolres12030025",
"author": "MB Skarzynska",
"doi-asserted-by": "publisher",
"first-page": "224",
"issue": "3",
"journal-title": "Audiol Res",
"key": "6541_CR28",
"unstructured": "Skarzynska MB, Matusiak M, Skarzynski PH (2022) Adverse audio-vestibular effects of drugs and vaccines used in the treatment and prevention of COVID-19: a review. Audiol Res 12(3):224–248. https://doi.org/10.3390/audiolres12030025",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1002/jmv.1890150110",
"author": "TN Kaul",
"doi-asserted-by": "publisher",
"first-page": "71",
"issue": "1",
"journal-title": "J Med Virol",
"key": "6541_CR29",
"unstructured": "Kaul TN, Middleton JE, Ogra PL (1985) Antiviral effect of flavonoids on human viruses. J Med Virol 15(1):71–79. https://doi.org/10.1002/jmv.1890150110",
"volume": "15",
"year": "1985"
},
{
"DOI": "10.1016/j.biopha.2021.111998",
"author": "X Shen",
"doi-asserted-by": "publisher",
"journal-title": "Biomed Pharmacother",
"key": "6541_CR30",
"unstructured": "Shen X, Yin F (2021) The mechanisms and clinical application of traditional Chinese medicine Lianhua-qingwen capsule. Biomed Pharmacother 142:111998. https://doi.org/10.1016/j.biopha.2021.111998",
"volume": "142",
"year": "2021"
},
{
"DOI": "10.1097/MD.0000000000020612",
"author": "H Chen",
"doi-asserted-by": "publisher",
"issue": "24",
"journal-title": "Medicine",
"key": "6541_CR31",
"unstructured": "Chen H, Song YP, Gao K, Zhao LT, Ma L (2020) Efficacy and safety of Jinhua Qinggan granules for coronavirus disease 2019 (COVID-19): a protocol of a systematic review and meta-analysis. Medicine 99(24):e20612. https://doi.org/10.1097/MD.0000000000020612",
"volume": "99",
"year": "2020"
},
{
"DOI": "10.1016/j.apsb.2020.06.009",
"author": "L Ni",
"doi-asserted-by": "publisher",
"first-page": "1149",
"issue": "7",
"journal-title": "Acta Pharm Sin B",
"key": "6541_CR32",
"unstructured": "Ni L, Chen L, Huang X, Han C, Xu J, Zhang H, Luan X, Zhao Y, Xu J, Yuan W et al (2020) Combating COVID-19 with integrated traditional Chinese and Western medicine in China. Acta Pharm Sin B 10(7):1149–1162. https://doi.org/10.1016/j.apsb.2020.06.009",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1097/MD.0000000000022715",
"author": "L Han",
"doi-asserted-by": "publisher",
"issue": "42",
"journal-title": "Medicine",
"key": "6541_CR33",
"unstructured": "Han L, Wang Y, Hu K, Tang Z, Song X (2020) The therapeutic efficacy of Huashi Baidu Formula combined with antiviral drugs in the treatment of COVID-19: a protocol for systematic review and meta-analysis. Medicine 99(42):e22715. https://doi.org/10.1097/MD.0000000000022715",
"volume": "99",
"year": "2020"
},
{
"DOI": "10.21037/apm-20-1478",
"doi-asserted-by": "publisher",
"key": "6541_CR34",
"unstructured": "Guo H, Zheng J, Huang G, Xiang Y, Lang C, Li B, Huang D, Sun Q, Luo Y, Zhang Y et al (2020) Xuebijing injection in the treatment of COVID-19: a retrospective case-control study. Ann Palliat Med. 9(5):3235248–3233248. https://doi.org/10.21037/apm-20-1478"
},
{
"DOI": "10.1002/jssc.202000898",
"doi-asserted-by": "publisher",
"key": "6541_CR35",
"unstructured": "Xie L, Wang Y, Yin H, Li J, Xu Z, Sun Z, Liu F, Zhang X, Liu S, Sun J et al (2021) Identification of the absorbed ingredients and metabolites in rats after an intravenous administration of Tanreqing injection using high‐performance liquid chromatography coupled with quadrupole time‐of‐flight mass spectrometry. J Sep Sci 44(10):2097–2112. https://doi.org/10.1002/jssc.202000898"
},
{
"DOI": "10.1186/s13020-020-00375-1",
"author": "H Luo",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Chin Med-UK",
"key": "6541_CR36",
"unstructured": "Luo H, Gao Y, Zou J, Zhang S, Chen H, Liu Q, Tan D, Han Y, Zhao Y, Wang S (2020) Reflections on treatment of COVID-19 with traditional Chinese medicine. Chin Med-UK 15(1):1–14. https://doi.org/10.1186/s13020-020-00375-1",
"volume": "15",
"year": "2020"
},
{
"DOI": "10.1016/j.biopha.2020.110500",
"author": "S Xin",
"doi-asserted-by": "publisher",
"first-page": "110500",
"journal-title": "Biomed & Pharmacother",
"key": "6541_CR37",
"unstructured": "Xin S, Cheng X, Zhu B, Liao X, Yang F, Song L, Shi Y, Guan X, Su R, Wang J (2020) Clinical retrospective study on the efficacy of Qingfei Paidu decoction combined with western medicine for COVID-19 treatment. Biomed & Pharmacother 129:110500. https://doi.org/10.1016/j.biopha.2020.110500",
"volume": "129",
"year": "2020"
},
{
"DOI": "10.1080/03639045.2020.1788070",
"author": "Q Tao",
"doi-asserted-by": "publisher",
"first-page": "1345",
"issue": "8",
"journal-title": "Drug Dev Ind Pharm",
"key": "6541_CR38",
"unstructured": "Tao Q, Du J, Li X, Zeng J, Tan B, Xu J, Lin W, Chen XL (2020) Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm 46(8):1345–1353. https://doi.org/10.1080/03639045.2020.1788070",
"volume": "46",
"year": "2020"
},
{
"DOI": "10.1016/S1875-5364(20)60038-3",
"author": "X Yan",
"doi-asserted-by": "publisher",
"first-page": "941",
"issue": "12",
"journal-title": "Chin J Nat Medicines",
"key": "6541_CR39",
"unstructured": "Yan X, Ying-Rong HUA, Shang J, Wei-Hong GE, Jun LIAO (2019) Traditional Chinese medicine network pharmacology study on exploring the mechanism of Xuebijing Injection in the treatment of coronavirus disease 2019. Chin J Nat Medicines 18(12):941–951. https://doi.org/10.1016/S1875-5364(20)60038-3",
"volume": "18",
"year": "2019"
},
{
"DOI": "10.1016/J.phrs.2020.104743",
"doi-asserted-by": "publisher",
"key": "6541_CR40",
"unstructured": "Ren J, Zhang AH, Wang XJ (2020) Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res 155:104743. https://doi.org/10.1016/J.phrs.2020.104743"
},
{
"DOI": "10.1039/C3FO60295H",
"author": "Y Hou",
"doi-asserted-by": "publisher",
"first-page": "1727",
"issue": "12",
"journal-title": "Food Funct",
"key": "6541_CR41",
"unstructured": "Hou Y, Jiang JG (2013) Origin and concept of medicine food homology and its application in modern functional foods. Food Funct 4(12):1727–1741. https://doi.org/10.1039/C3FO60295H",
"volume": "4",
"year": "2013"
},
{
"DOI": "10.1186/1758-2946-6-13",
"author": "J Ru",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "J Cheminform",
"key": "6541_CR42",
"unstructured": "Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y et al (2014) TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 6:1–6. https://doi.org/10.1186/1758-2946-6-13",
"volume": "6",
"year": "2014"
},
{
"DOI": "10.1021/mp100465q",
"author": "L Chen",
"doi-asserted-by": "publisher",
"first-page": "889",
"issue": "3",
"journal-title": "Mol Pharm",
"key": "6541_CR43",
"unstructured": "Chen L, Li Y, Zhao Q et al (2011) ADME evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive Bayesian classification techniques. Mol Pharm 8(3):889–900",
"volume": "8",
"year": "2011"
},
{
"DOI": "10.1016/j.abb.2015.05.011",
"author": "NM Cerqueira",
"doi-asserted-by": "publisher",
"first-page": "56",
"journal-title": "Arch Biochem Biophys",
"key": "6541_CR44",
"unstructured": "Cerqueira NM, Gesto D, Oliveira EF, Santos-Martins D, Brás NF, Sousa SF, Fernandes PA, Ramos MJ (2015) Receptor-based virtual screening protocol for drug discovery. Arch Biochem Biophys 582:56–67. https://doi.org/10.1016/j.abb.2015.05.011",
"volume": "582",
"year": "2015"
},
{
"DOI": "10.1016/j.jmgm.2014.11.015",
"author": "MA Islam",
"doi-asserted-by": "publisher",
"first-page": "20",
"issue": "2",
"journal-title": "J Mol Graph Model",
"key": "6541_CR45",
"unstructured": "Islam MA, Pillay TS (2015) Exploration of the structural requirements of HIV-protease inhibitors using pharmacophore virtual screening and molecular docking approaches for lead identification. J Mol Graph Model 56(2):20–30. https://doi.org/10.1016/j.jmgm.2014.11.015",
"volume": "56",
"year": "2015"
},
{
"DOI": "10.1039/C5MB00767D",
"author": "MA Islam",
"doi-asserted-by": "publisher",
"first-page": "982",
"issue": "3",
"journal-title": "Mol Biosyst",
"key": "6541_CR46",
"unstructured": "Islam MA, Pillay TS (2016) Structural requirements for potential HIV-integrase inhibitors identified using pharmacophore-based virtual screening and molecular dynamics studies. Mol Biosyst 12(3):982–993. https://doi.org/10.1039/C5MB00767D",
"volume": "12",
"year": "2016"
},
{
"DOI": "10.1016/j.jmgm.2017.10.002",
"author": "W Du",
"doi-asserted-by": "publisher",
"first-page": "96",
"journal-title": "J Mol Graph Model",
"key": "6541_CR47",
"unstructured": "Du W, Zuo K, Sun X, Liu W, Yan X, Liang L, Wan H, Chen F, Hu J (2017) An effective HIV-1 integrase inhibitor screening platform: rationality validation of drug screening conformational mobility and molecular recognition analysis for PFV integrase complex with viral DNA. J Mol Graph Model 78:96–109. https://doi.org/10.1016/j.jmgm.2017.10.002",
"volume": "78",
"year": "2017"
},
{
"DOI": "10.1007/BF00124324",
"author": "G Jones",
"doi-asserted-by": "publisher",
"first-page": "532",
"issue": "6",
"journal-title": "J Comput Aided Mol Des",
"key": "6541_CR48",
"unstructured": "Jones G, Willett P, Glen RC (1995) A genetic algorithm for flexible molecular overlay and pharmacophore elucidation. J Comput Aided Mol Des 9(6):532–549. https://doi.org/10.1007/BF00124324",
"volume": "9",
"year": "1995"
},
{
"DOI": "10.1002/jcc.21334",
"author": "O Trott",
"doi-asserted-by": "publisher",
"first-page": "455",
"journal-title": "J Comput Chem",
"key": "6541_CR49",
"unstructured": "Trott O, Olson AJ (2010) Autodock vina: improving the speed and accuracy of docking with a new scoring function efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1038/srep15290",
"author": "J Li",
"doi-asserted-by": "publisher",
"journal-title": "Sci Rep",
"key": "6541_CR50",
"unstructured": "Li J, Zhao P, Li Y, Tian Y, Wang Y (2015) Systems pharmacology-based dissection of mechanisms of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci Rep 5:15290. https://doi.org/10.1038/srep15290",
"volume": "5",
"year": "2015"
},
{
"DOI": "10.3390/ijms13066964",
"author": "X Xu",
"doi-asserted-by": "publisher",
"first-page": "6964",
"issue": "6",
"journal-title": "Int J Mol Sci",
"key": "6541_CR51",
"unstructured": "Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, Duan J, Yang L (2012) A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 13(6):6964–6982. https://doi.org/10.3390/ijms13066964",
"volume": "13",
"year": "2012"
},
{
"DOI": "10.1016/j.jep.2012.09.051",
"author": "W Tao",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "J Ethnopharmacol",
"key": "6541_CR52",
"unstructured": "Tao W, Xu X, Wang X, Li B, Wang Y, Li Y, Yang L (2013) Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal radix curcumae formula for application to cardiovascular disease. J Ethnopharmacol 145(1):1–10. https://doi.org/10.1016/j.jep.2012.09.051",
"volume": "145",
"year": "2013"
},
{
"DOI": "10.1021/acs.jcim.9b00445",
"author": "R Kong",
"doi-asserted-by": "publisher",
"first-page": "3556",
"issue": "8",
"journal-title": "J Chem Inf Model",
"key": "6541_CR53",
"unstructured": "Kong R, Wang F, Zhang J, Wang F, Chang S (2019) CoDockPP: a multistage approach for global and site-specific protein–protein docking. J Chem Inf Model 59(8):3556–3564. https://doi.org/10.1021/acs.jcim.9b00445",
"volume": "59",
"year": "2019"
},
{
"DOI": "10.1002/jcc.20035",
"author": "J Wang",
"doi-asserted-by": "publisher",
"first-page": "1157",
"journal-title": "J Comput Chem",
"key": "6541_CR54",
"unstructured": "Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1063/1.445869",
"author": "WL Jorgensen",
"doi-asserted-by": "publisher",
"first-page": "926",
"issue": "2",
"journal-title": "J Chem Phys",
"key": "6541_CR55",
"unstructured": "Jorgensen WL, Chandrasekhar J, Madura JD (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935. https://doi.org/10.1063/1.445869",
"volume": "79",
"year": "1983"
},
{
"DOI": "10.1002/jcc.540130805",
"author": "S Miyamoto",
"doi-asserted-by": "publisher",
"first-page": "952",
"issue": "8",
"journal-title": "J Comput Chem",
"key": "6541_CR56",
"unstructured": "Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962. https://doi.org/10.1002/jcc.540130805",
"volume": "13",
"year": "1992"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"author": "W Humphrey",
"doi-asserted-by": "publisher",
"first-page": "33",
"issue": "1",
"journal-title": "J Mol Graph",
"key": "6541_CR57",
"unstructured": "Humphrey W, Dalke A, Schulten K (1996) VMD-Visual molecular dynamics. J Mol Graph 14(1):33–38. https://doi.org/10.1016/0263-7855(96)00018-5",
"volume": "14",
"year": "1996"
},
{
"DOI": "10.1371/journal.pone.0076045",
"author": "H Wan",
"doi-asserted-by": "publisher",
"issue": "10",
"journal-title": "PLoS ONE",
"key": "6541_CR58",
"unstructured": "Wan H, Hu J, Li K, Tian XH, Chang S (2013) Molecular dynamics simulations of DNA-free and DNA-bound TAL effectors. PLoS ONE 8(10):e76045. https://doi.org/10.1371/journal.pone.0076045",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.1080/08927022.2019.1579327",
"author": "H Duan",
"doi-asserted-by": "publisher",
"first-page": "694",
"issue": "9",
"journal-title": "Mol Simul",
"key": "6541_CR59",
"unstructured": "Duan H, Liu X, Zhuo W, Meng J, Gu J, Sun X, Zuo K, Luo Q, Luo Y, Tang D et al (2019) 3D-QSAR and molecular recognition of Klebsiella pneumoniae NDM-1 inhibitors. Mol Simul 45(9):694–705. https://doi.org/10.1080/08927022.2019.1579327",
"volume": "45",
"year": "2019"
},
{
"DOI": "10.1016/j.jmgm.2003.12.005",
"author": "M Feig",
"doi-asserted-by": "publisher",
"first-page": "377",
"issue": "5",
"journal-title": "J Mol Graph Model",
"key": "6541_CR60",
"unstructured": "Feig M, Karanicolas J, Brooks IIICL (2004) MMTSB tool set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model 22(5):377–395. https://doi.org/10.1016/j.jmgm.2003.12.005",
"volume": "22",
"year": "2004"
},
{
"DOI": "10.1021/ar000033j",
"author": "PA Kollman",
"doi-asserted-by": "publisher",
"first-page": "889",
"issue": "12",
"journal-title": "Acc Chem Res",
"key": "6541_CR61",
"unstructured": "Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897. https://doi.org/10.1021/ar000033j",
"volume": "33",
"year": "2000"
},
{
"DOI": "10.1016/S0959-440X(00)00197-4",
"author": "T Simonson",
"doi-asserted-by": "publisher",
"first-page": "243",
"issue": "2",
"journal-title": "Curr Opin Struct Biol",
"key": "6541_CR62",
"unstructured": "Simonson T (2001) Macromolecular electrostatics: continuum models and their growing pains. Curr Opin Struct Biol 11(2):243–252. https://doi.org/10.1016/S0959-440X(00)00197-4",
"volume": "11",
"year": "2001"
},
{
"DOI": "10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A",
"author": "J Welser",
"doi-asserted-by": "publisher",
"first-page": "217",
"issue": "2",
"journal-title": "J Comput Chem",
"key": "6541_CR63",
"unstructured": "Welser J, Shenkin PS, Still WC (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20(2):217–230. https://doi.org/10.1002/(SICI)1096-987X(19990130)20:2%3c217::AID-JCC4%3e3.0.CO;2-A",
"volume": "20",
"year": "1999"
},
{
"DOI": "10.1016/j.phymed.2022.154481",
"author": "Y Lin",
"doi-asserted-by": "publisher",
"journal-title": "Phytomedicine",
"key": "6541_CR64",
"unstructured": "Lin Y, Zhang Y, Wang D, Yang B, Shen YQ (2022) Computer especially AI-assisted drug virtual screening and design in traditional Chinese medicine. Phytomedicine 107:154481. https://doi.org/10.1016/j.phymed.2022.154481",
"volume": "107",
"year": "2022"
},
{
"DOI": "10.1021/ci200617d",
"author": "A Hamza",
"doi-asserted-by": "publisher",
"first-page": "963",
"issue": "4",
"journal-title": "J Chem Inf Model",
"key": "6541_CR65",
"unstructured": "Hamza A, Wei NN, Zhan CG (2012) Ligand-based virtual screening approach using a new scoring function. J Chem Inf Model 52(4):963–974. https://doi.org/10.1021/ci200617d",
"volume": "52",
"year": "2012"
},
{
"DOI": "10.3390/ijms18050761",
"author": "K Zuo",
"doi-asserted-by": "publisher",
"issue": "5",
"journal-title": "Int J Mol Sci",
"key": "6541_CR66",
"unstructured": "Zuo K, Liang L, Du W, Sun X, Liu W, Gou X, Wan H, Hu J (2017) 3D-QSAR molecular docking and molecular dynamics simulation of Pseudomonas aeruginosa LpxC inhibitors. Int J Mol Sci 18(5):761. https://doi.org/10.3390/ijms18050761",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1186/s43556-022-00074-3",
"author": "D Guo",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Mol Biomed",
"key": "6541_CR67",
"unstructured": "Guo D, Duan H, Cheng Y, Wang Y, Hu J, Shi H (2022) Omicron-included mutation-induced changes in epitopes of SARS-CoV-2 spike protein and effectiveness assessments of current antibodies. Mol Biomed 3(1):1–18. https://doi.org/10.1186/s43556-022-00074-3",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1016/j.jep.2019.112044",
"author": "L Ren",
"doi-asserted-by": "publisher",
"journal-title": "J Ethnopharmacol",
"key": "6541_CR68",
"unstructured": "Ren L, Zheng X, Liu J, Li W, Fu W, Tang Q, Wang J, Du G (2019) Network pharmacology study of traditional Chinese medicines for stroke treatment and effective constituents screening. J Ethnopharmacol 242:112044. https://doi.org/10.1016/j.jep.2019.112044",
"volume": "242",
"year": "2019"
},
{
"DOI": "10.1016/j.isci.2021.102148",
"author": "Z Li",
"doi-asserted-by": "publisher",
"issue": "3",
"journal-title": "iScience",
"key": "6541_CR69",
"unstructured": "Li Z, Yao Y, Cheng X, Chen Q, Zhao W, Ma S, Li Z, Zhou H, Li W, Fei T (2021) A computational framework of host-based drug repositioning for broad-spectrum antivirals against RNA viruses. iScience 24(3):102148. https://doi.org/10.1016/j.isci.2021.102148",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1002/minf.202100002",
"author": "X Li",
"doi-asserted-by": "publisher",
"first-page": "2100002",
"issue": "3",
"journal-title": "Mol Inform",
"key": "6541_CR70",
"unstructured": "Li X, Jiang Q, Yang X (2022) Discovery of inhibitors for mycobacterium tuberculosis peptide deformylase based on virtual screening in silico. Mol Inform 41(3):2100002. https://doi.org/10.1002/minf.202100002",
"volume": "41",
"year": "2022"
},
{
"DOI": "10.1080/14756366.2021.1954919",
"author": "M Pavan",
"doi-asserted-by": "publisher",
"first-page": "1645",
"issue": "1",
"journal-title": "J Enzym Inhib Med Chem",
"key": "6541_CR71",
"unstructured": "Pavan M, Bolcato G, Bassani D, Sturlese M, Moro S (2021) Supervised molecular dynamics (SuMD) insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J Enzym Inhib Med Chem 36(1):1645–1649. https://doi.org/10.1080/14756366.2021.1954919",
"volume": "36",
"year": "2021"
},
{
"DOI": "10.1016/j.jep.2013.02.004",
"author": "H Liu",
"doi-asserted-by": "publisher",
"first-page": "773",
"issue": "3",
"journal-title": "J Ethnopharmacol",
"key": "6541_CR72",
"unstructured": "Liu H, Wang J, Zhou W, Wang Y, Yang L (2013) Systems approaches and polypharmacology for drug discovery from herbal medicines: an example using licorice. J Ethnopharmacol 146(3):773–793. https://doi.org/10.1016/j.jep.2013.02.004",
"volume": "146",
"year": "2013"
},
{
"DOI": "10.1111/cpr.12949",
"author": "QD Xia",
"doi-asserted-by": "publisher",
"first-page": "e12949",
"issue": "12",
"journal-title": "Cell Prolif",
"key": "6541_CR73",
"unstructured": "Xia QD, Xun Y, Lu JL, Lu YC, Yang YY, Zhou P, Hu J, Li C, Wang SG (2020) Network pharmacology and molecular docking analyses on Lianhua Qingwen capsule indicate Akt1 is a potential target to treat and prevent COVID-19. Cell Prolif 53(12):e12949. https://doi.org/10.1111/cpr.12949",
"volume": "53",
"year": "2020"
},
{
"DOI": "10.1007/s11655-020-3476-x",
"author": "L Gao",
"doi-asserted-by": "publisher",
"first-page": "527",
"journal-title": "Chin J Integr Med",
"key": "6541_CR74",
"unstructured": "Gao L, Xu J, Chen S (2020) In silico screening of potential Chinese herbal medicine against COVID-19 by targeting SARS-CoV-2 3CLpro and angiotensin converting enzyme II using molecular docking. Chin J Integr Med 26:527–532. https://doi.org/10.1007/s11655-020-3476-x",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1097/MD.0000000000020531",
"author": "Q Zhang",
"doi-asserted-by": "publisher",
"issue": "24",
"journal-title": "Medicine (Baltimore)",
"key": "6541_CR75",
"unstructured": "Zhang Q, Cao F, Wang Y, Xu X, Sun Y, Li J, Qi X, Sun S, Ji G, Song B (2020) The efficacy and safety of Jinhua Qinggan granule (JHQG) in the treatment of coronavirus disease 2019 (COVID-19): a protocol for systematic review and meta analysis. Medicine (Baltimore) 99(24):e20531. https://doi.org/10.1097/MD.0000000000020531",
"volume": "99",
"year": "2020"
},
{
"DOI": "10.1155/2015/731765",
"author": "W Jia",
"doi-asserted-by": "publisher",
"first-page": "731765",
"issue": "1",
"journal-title": "Sci World J",
"key": "6541_CR76",
"unstructured": "Jia W, Wang C, Wang Y, Pan G, Jiang M, Li Z, Zhu Y (2015) Qualitative and quantitative analysis of the major constituents in Chinese medical preparation Lianhua-Qingwen capsule by UPLC-DAD-QTOF-MS. Sci World J 2015(1):731765. https://doi.org/10.1155/2015/731765",
"volume": "2015",
"year": "2015"
},
{
"DOI": "10.1016/j.jep.2021.113871",
"author": "S Jia",
"doi-asserted-by": "publisher",
"journal-title": "J Ethnopharmacol",
"key": "6541_CR77",
"unstructured": "Jia S, Luo H, Liu X, Fan X, Huang Z, Lu S, Shen L, Guo S, Liu Y, Wang Z et al (2021) Dissecting the novel mechanism of Reduning injection in treating Coronavirus Disease 2019 (COVID-19) based on network pharmacology and experimental verification. J Ethnopharmacol 273:113871. https://doi.org/10.1016/j.jep.2021.113871",
"volume": "273",
"year": "2021"
},
{
"DOI": "10.1016/S1875-5364(16)30091-7",
"author": "F Zhang",
"doi-asserted-by": "publisher",
"first-page": "769",
"issue": "10",
"journal-title": "Chin J Nat Medicines",
"key": "6541_CR78",
"unstructured": "Zhang F, Liang SUN, Shou-Hong GAO, Wan-Sheng CHEN, Yi-Feng CHAI (2016) LC-MS/MS analysis and pharmacokinetic study on five bioactive constituents of Tanreqing injection in rats. Chin J Nat Medicines 14(10):769–775. https://doi.org/10.1016/S1875-5364(16)30091-7",
"volume": "14",
"year": "2016"
},
{
"DOI": "10.1186/s13020-021-00427-0",
"author": "S Xia",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Chin Med-UK",
"key": "6541_CR79",
"unstructured": "Xia S, Zhong Z, Gao B, Vong CT, Lin X, Cai J, Gao H, Chan G, Li C (2021) The important herbal pair for the treatment of COVID-19 and its possible mechanisms. Chin Med-UK 16(1):1–16. https://doi.org/10.1186/s13020-021-00427-0",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1016/j.jsps.2020.08.002",
"author": "T He",
"doi-asserted-by": "publisher",
"first-page": "1138",
"issue": "9",
"journal-title": "Saudi Pharm J",
"key": "6541_CR80",
"unstructured": "He T, Qu R, Qin C, Wang Z, Zhang Y, Shao X, Lu T (2020) Potential mechanisms of Chinese herbal medicine that implicated in the treatment of COVID-19 related renal injury. Saudi Pharm J 28(9):1138–1148",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1007/s11096-020-01153-7",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "35",
"issue": "1",
"journal-title": "Int J Clin Pharm-Net",
"key": "6541_CR81",
"unstructured": "Wang C, Sun S, Ding X (2021) The therapeutic effects of traditional Chinese medicine on COVID-19: a narrative review. Int J Clin Pharm-Net 43(1):35–45. https://doi.org/10.1007/s11096-020-01153-7",
"volume": "43",
"year": "2021"
},
{
"DOI": "10.7150/ijms.53685",
"author": "X Zhang",
"doi-asserted-by": "publisher",
"first-page": "1866",
"issue": "8",
"journal-title": "Int J Med Sci",
"key": "6541_CR82",
"unstructured": "Zhang X, Gao R, Zhou Z, Tang X, Lin J, Wang L, Zhou X, Shen TA (2021) Network pharmacology based approach for predicting active ingredients and potential mechanism of Lianhuaqingwen capsule in treating COVID-19. Int J Med Sci 18(8):1866. https://doi.org/10.7150/ijms.53685",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1007/s11655-022-3686-5",
"author": "Z Xiao",
"doi-asserted-by": "publisher",
"first-page": "205",
"issue": "3",
"journal-title": "Chin J Integr Med",
"key": "6541_CR83",
"unstructured": "Xiao Z, Xu H, Qu Z, Ma XY, Huang BX, Sun MS, Wang BQ, Wang GY (2023) Active ingredients of Reduning injections maintain high potency against SARS-CoV-2 variants. Chin J Integr Med 29(3):205–212. https://doi.org/10.1007/s11655-022-3686-5",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.1016/j.phymed.2020.153401",
"author": "M Ye",
"doi-asserted-by": "publisher",
"journal-title": "Phytomedicine",
"key": "6541_CR84",
"unstructured": "Ye M, Luo G, Ye D, She M, Sun N, Lu YJ, Zheng J (2021) Network pharmacology molecular docking integrated surface plasmon resonance technology reveals the mechanism of Toujie quwen granules against coronavirus disease 2019 pneumonia. Phytomedicine 85:153401. https://doi.org/10.1016/j.phymed.2020.153401",
"volume": "85",
"year": "2021"
},
{
"DOI": "10.1038/s41598-020-69337-9",
"author": "R Yoshino",
"doi-asserted-by": "publisher",
"issue": "1",
"journal-title": "Sci Rep",
"key": "6541_CR85",
"unstructured": "Yoshino R, Yasuo N, Sekijima M (2020) Identification of key interactions between SARS-CoV-2 main protease and inhibitor drug candidates. Sci Rep 10(1):12493. https://doi.org/10.1038/s41598-020-69337-9",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.bpc.2007.09.008",
"author": "JP Hu",
"doi-asserted-by": "publisher",
"first-page": "69",
"issue": "2–3",
"journal-title": "Biophys Chem",
"key": "6541_CR86",
"unstructured": "Hu JP, Gong XQ, Su JG, Zu Chen W, Wang CX (2008) Study on the molecular mechanism of inhibiting HIV-1 integrase by EBR28 peptide via molecular modeling approach. Biophys Chem 132(2–3):69–80. https://doi.org/10.1016/j.bpc.2007.09.008",
"volume": "132",
"year": "2008"
},
{
"DOI": "10.1021/ci200227u",
"author": "RA Laskowski",
"doi-asserted-by": "publisher",
"journal-title": "J Chem Inf Model",
"key": "6541_CR87",
"unstructured": "Laskowski RA, Swindells MB (2011) Ligplot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. https://doi.org/10.1021/ci200227u",
"year": "2011"
},
{
"DOI": "10.1016/j.compbiomed.2022.105468",
"author": "A Parihar",
"doi-asserted-by": "publisher",
"journal-title": "Comput Biol Med",
"key": "6541_CR88",
"unstructured": "Parihar A, Sonia ZF, Akter F, Ali MA, Hakim FT, Hossain MS (2022) Phytochemicals-based targeting RdRp and main protease of SARS-CoV-2 using docking and steered molecular dynamic simulation: a promising therapeutic approach for tackling COVID-19. Comput Biol Med 145:105468. https://doi.org/10.1016/j.compbiomed.2022.105468",
"volume": "145",
"year": "2022"
},
{
"DOI": "10.1103/PhysRevLett.88.138101",
"author": "M Tarek",
"doi-asserted-by": "publisher",
"issue": "13",
"journal-title": "Phys Rev Lett",
"key": "6541_CR89",
"unstructured": "Tarek M, Tobias DJ (2002) Role of protein-water hydrogen bond dynamics in the protein dynamical transition. Phys Rev Lett 88(13):138101. https://doi.org/10.1103/PhysRevLett.88.138101",
"volume": "88",
"year": "2002"
},
{
"DOI": "10.1038/s41467-020-16954-7",
"author": "DW Kneller",
"doi-asserted-by": "publisher",
"first-page": "3202",
"issue": "1",
"journal-title": "Nat Commun",
"key": "6541_CR90",
"unstructured": "Kneller DW, Phillips G, O’Neill HM, Jedrzejczak R, Stols L, Langan P, Joachimiak A, Coates L, Kovalevsky A (2020) Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat Commun 11(1):3202. https://doi.org/10.1038/s41467-020-16954-7",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3390/ijms22137048",
"author": "F Mangiavacchi",
"doi-asserted-by": "publisher",
"issue": "13",
"journal-title": "Int J Mol Sci",
"key": "6541_CR91",
"unstructured": "Mangiavacchi F, Botwina P, Menichetti E, Bagnoli L, Rosati O, Marini F, Fonseca SF, Abenante L, Alves D, Dabrowska A et al (2021) Seleno-functionalization of quercetin improves the non-covalent inhibition of Mpro and its antiviral activity in cells against SARS-CoV-2. Int J Mol Sci 22(13):7048. https://doi.org/10.3390/ijms22137048",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.3389/fphar.2021.705252",
"author": "B Zhou",
"doi-asserted-by": "publisher",
"journal-title": "Front Pharmacol",
"key": "6541_CR92",
"unstructured": "Zhou B, Yuan Y, Shi L, Hu S, Wang D, Yang Y, Pan Y, Kong D, Shikov AN, Duez P et al (2021) Creation of an anti-inflammatory leptin-dependent anti-obesity celastrol mimic with better druggability. Front Pharmacol 12:705252. https://doi.org/10.3389/fphar.2021.705252",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1002/2211-5463.12875",
"author": "M Tsuji",
"doi-asserted-by": "publisher",
"first-page": "995",
"issue": "6",
"journal-title": "FEBS Open Bio",
"key": "6541_CR93",
"unstructured": "Tsuji M (2020) Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio 10(6):995–1004",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkac956",
"author": "S Kim",
"doi-asserted-by": "publisher",
"first-page": "D1373",
"issue": "D1",
"journal-title": "Nucleic Acids Res",
"key": "6541_CR94",
"unstructured": "Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B et al (2023) PubChem 2023 update. Nucleic Acids Res 51(D1):D1373–D1380. https://doi.org/10.1093/nar/gkac956",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1186/1758-2946-3-33",
"author": "NM O'Boyle",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "J Cheminform",
"key": "6541_CR95",
"unstructured": "O’Boyle NM, Banck M, James CA et al (2011) Open babel: an open chemical toolbox. J Cheminform 3(1):1–14. https://doi.org/10.1186/1758-2946-3-33",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.1002/jcc.20084",
"author": "EF Pettersen",
"doi-asserted-by": "publisher",
"first-page": "1605",
"issue": "13",
"journal-title": "J Comput Chem",
"key": "6541_CR96",
"unstructured": "Pettersen EF, Goddard TD, Huang CC, Morley C, Vandermeersch T, Hutchison GR (2004) UCSF chimera-a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1016/j.taap.2022.115922",
"author": "D Hoer",
"doi-asserted-by": "publisher",
"journal-title": "Toxicol Appl Pharm",
"key": "6541_CR97",
"unstructured": "Hoer D, Barton HA, Paini A, Bartels M, Ingle B, Domoradzki J, Fisher J, Embry M, Villanueva P, Miller D et al (2022) Predicting nonlinear relationships between external and internal concentrations with physiologically based pharmacokinetic modeling. Toxicol Appl Pharm 440:115922. https://doi.org/10.1016/j.taap.2022.115922",
"volume": "440",
"year": "2022"
},
{
"DOI": "10.1186/s13321-018-0283-x",
"author": "J Dong",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "J cheminformatics",
"key": "6541_CR98",
"unstructured": "Dong J, Wang NN, Yao ZJ, Zhang L, Cheng Y, Ouyang D, Lu AP, Cao DS (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J cheminformatics 10:1–11. https://doi.org/10.1186/s13321-018-0283-x",
"volume": "10",
"year": "2018"
},
{
"DOI": "10.1038/srep42717",
"author": "A Daina",
"doi-asserted-by": "publisher",
"issue": "1",
"journal-title": "Sci Rep",
"key": "6541_CR99",
"unstructured": "Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):42717. https://doi.org/10.1038/srep42717",
"volume": "7",
"year": "2017"
},
{
"DOI": "10.1021/acs.jcim.8b00677",
"doi-asserted-by": "publisher",
"key": "6541_CR100",
"unstructured": "Stork C, Chen Y, Sicho M, Kirchmair J (2019) Hit Dexter 2.0: machine-learning models for the prediction of frequent hitters. J Chem Inf Model 59(3):1030–1043.https://doi.org/10.1021/acs.jcim.8b00677"
},
{
"DOI": "10.1080/07391102.2021.1872419",
"author": "S Shah",
"doi-asserted-by": "publisher",
"first-page": "5643",
"issue": "12",
"journal-title": "J Biomol Struct Dyn",
"key": "6541_CR101",
"unstructured": "Shah S, Chaple D, Arora S, Yende S, Mehta C, Nayak U (2022) Prospecting for cressa cretica to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2. J Biomol Struct Dyn 40(12):5643–5652. https://doi.org/10.1080/07391102.2021.1872419",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.3389/fchem.2021.692168",
"author": "JC Ferreira",
"doi-asserted-by": "publisher",
"journal-title": "Front Chem",
"key": "6541_CR102",
"unstructured": "Ferreira JC, Fadl S, Villanueva AJ, Rabeh WM (2021) Catalytic dyad residues His41 and Cys145 impact the catalytic activity and overall conformational fold of the main SARS-CoV-2 protease 3-chymotrypsin-like protease. Front Chem 9:692168. https://doi.org/10.3389/fchem.2021.692168",
"volume": "9",
"year": "2021"
}
],
"reference-count": 102,
"references-count": 102,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s00894-025-06541-2"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Feasibility of the inhibitor development for SARS-CoV-2: a systematic approach for drug design",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "31"
}