Computational identification of selected bioactive compounds from Cedrus deodara as inhibitors against SARS-CoV-2 main protease: a pharmacoinformatics study
et al., Indian Drugs, doi:10.53879/id.61.02.13859, Feb 2024
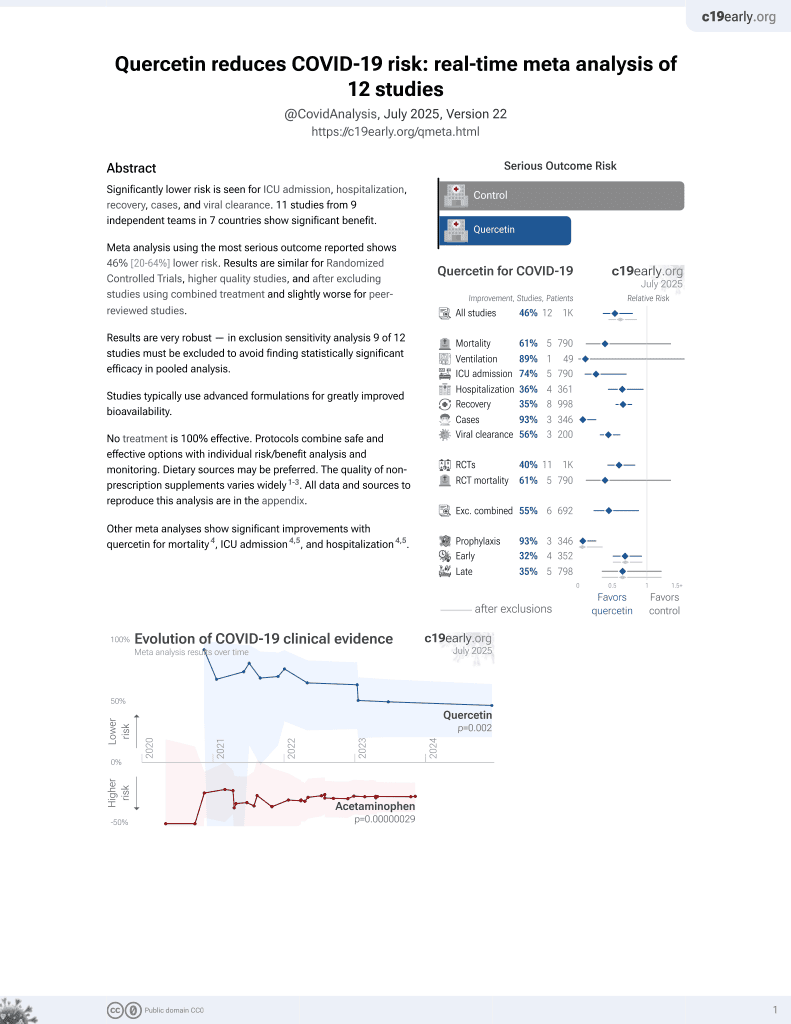
Quercetin for COVID-19
27th treatment shown to reduce risk in
July 2021, now with p = 0.002 from 12 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
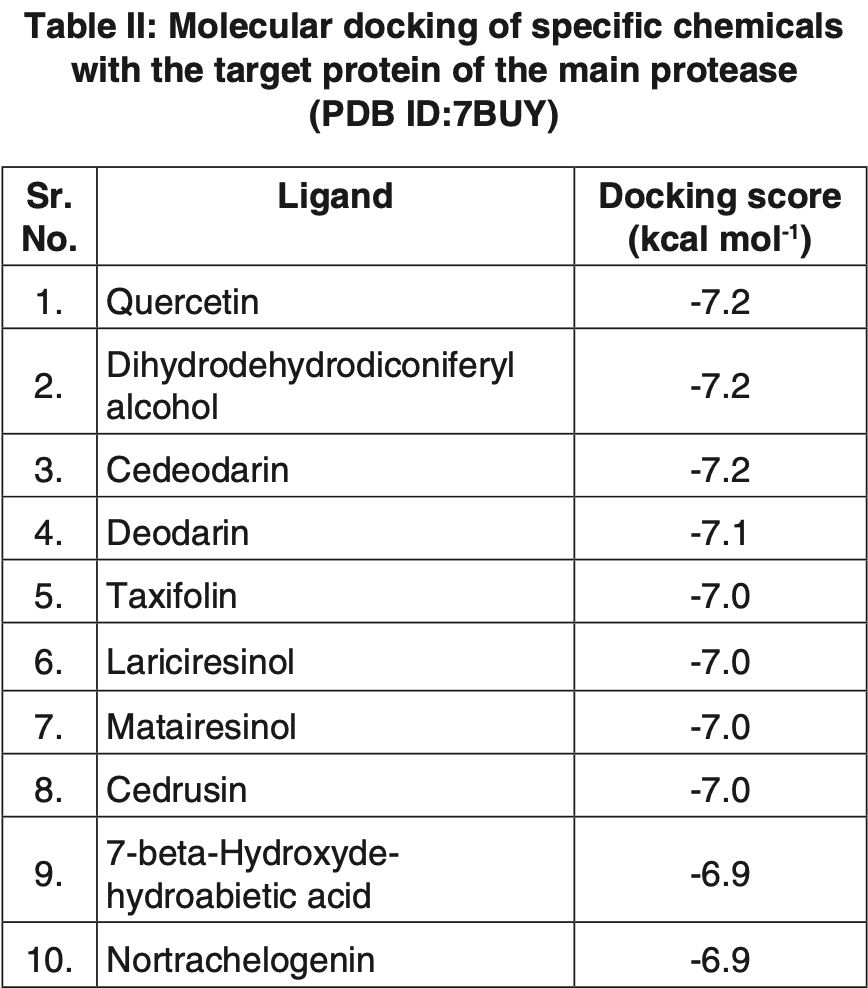
In silico study showing that quercetin binds strongly to the SARS-CoV-2 main protease (Mpro). Authors found that out of 49 phytoconstituents from Cedrus deodara, quercetin exhibited the lowest binding energy of approximately -7.2 kcal/mol when interacting with Mpro, forming 4 hydrogen bonds and 1 hydrophobic interaction. Molecular dynamics simulations confirmed the stability of the quercetin-Mpro complex over a 50ns simulation period.
91 preclinical studies support the efficacy of quercetin for COVID-19:
In silico studies predict inhibition of SARS-CoV-2, or minimization of side effects, with quercetin or metabolites via binding to the spikeA,11,12,18,19,32,34,35,37,40,48,49,51,52,75 (and specifically the receptor binding domainB,8), MproC,7,8,11,12,16,18,20,22,24,26,28,30,33,34,37,40,44,46-48,52-55,72 , RNA-dependent RNA polymeraseD,8,10-12,18,42 , PLproE,12,47,55 , ACE2F,27,32,33,37,38,47,51 , TMPRSS2G,32, nucleocapsidH,12, helicaseI,12,39,44 , endoribonucleaseJ,49, NSP16/10K,15, cathepsin LL,36, Wnt-3M,32, FZDN,32, LRP6O,32, ezrinP,50, ADRPQ,48, NRP1R,51, EP300S,25, PTGS2T,33, HSP90AA1U,25,33 , matrix metalloproteinase 9V,41, IL-6W,31,45 , IL-10X,31, VEGFAY,45, and RELAZ,45 proteins, and inhibition of spike-ACE2 interactionAA,9.
In vitro studies demonstrate inhibition of the MproC,24,58,63,71 protein, and inhibition of spike-ACE2 interactionAA,59.
In vitro studies demonstrate efficacy in Calu-3AB,62, A549AC,31, HEK293-ACE2+AD,70, Huh-7AE,35, Caco-2AF,61, Vero E6AG,29,52,61 , mTECAH,64, RAW264.7AI,64, and HLMECAJ,9 cells.
Animal studies demonstrate efficacy in K18-hACE2 miceAK,67, db/db miceAL,64,74 , BALB/c miceAM,73, and rats29.
Quercetin reduced proinflammatory cytokines and protected lung and kidney tissue against LPS-induced damage in mice73, inhibits LPS-induced cytokine storm by modulating key inflammatory and antioxidant pathways in macrophages14, may block ACE2-spike interaction and NLRP3 inflammasome, limiting viral entry and inflammation5, upregulates the SIRT1/AMPK axis to inhibit oxidative injury and accelerate viral clearance76, inhibits SARS-CoV-2 ORF3a ion channel activity, which contributes to viral pathogenicity and cytotoxicity66, may alleviate COVID-19 ARDS via inhibition of EGFR and JAK2 inflammatory targets1, and may destabilize the Spike protein, IL-6R, and integrins via conserved residues, blocking viral entry, hyperinflammation, and platelet aggregation77.
1.
Gupta et al., Harnessing phytoconstituents to treat COVID-19 triggered acute respiratory distress syndrome: Insights from network pharmacology, and molecular modeling, Phytochemistry Letters, doi:10.1016/j.phytol.2025.104105.
2.
Sun et al., Feasibility of the inhibitor development for SARS-CoV-2: a systematic approach for drug design, Journal of Molecular Modeling, doi:10.1007/s00894-025-06541-2.
3.
Torabfam et al., Improving quercetin solubility via structural modification enhances dual-target coronavirus entry: an integrated in-vitro and in-silico study, Scientific Reports, doi:10.1038/s41598-025-27374-2.
4.
Abdelhameed et al., Phytochemical and antiviral investigation of Cynanchum acutum L. extract and derived semi-synthetic analogs targeting SARS-CoV-2 main protease, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-025-00907-2.
5.
Manikyam et al., INP-Guided Network Pharmacology Discloses Multi-Target Therapeutic Strategy Against Cytokine and IgE Storms in the SARS-CoV-2 NB.1.8.1 Variant, Research Square, doi:10.21203/rs.3.rs-6819274/v1.
6.
Makoana et al., Integration of metabolomics and chemometrics with in-silico and in-vitro approaches to unravel SARS-Cov-2 inhibitors from South African plants, PLOS ONE, doi:10.1371/journal.pone.0320415.
7.
Bano et al., Biochemical Screening of Phytochemicals and Identification of Scopoletin as a Potential Inhibitor of SARS-CoV-2 Mpro, Revealing Its Biophysical Impact on Structural Stability, Viruses, doi:10.3390/v17030402.
8.
Rajamanickam et al., Exploring the Potential of Siddha Formulation MilagaiKudineer-Derived Phytotherapeutics Against SARS-CoV-2: An In-Silico Investigation for Antiviral Intervention, Journal of Pharmacy and Pharmacology Research, doi:10.26502/fjppr.0105.
9.
Moharram et al., Secondary metabolites of Alternaria alternate appraisal of their SARS-CoV-2 inhibitory and anti-inflammatory potentials, PLOS ONE, doi:10.1371/journal.pone.0313616.
10.
Metwaly et al., Integrated study of Quercetin as a potent SARS-CoV-2 RdRp inhibitor: Binding interactions, MD simulations, and In vitro assays, PLOS ONE, doi:10.1371/journal.pone.0312866.
11.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
12.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
13.
Pan et al., Decoding the mechanism of Qingjie formula in the prevention of COVID-19 based on network pharmacology and molecular docking, Heliyon, doi:10.1016/j.heliyon.2024.e39167.
14.
Xu et al., Quercetin inhibited LPS-induced cytokine storm by interacting with the AKT1-FoxO1 and Keap1-Nrf2 signaling pathway in macrophages, Scientific Reports, doi:10.1038/s41598-024-71569-y.
15.
Tamil Selvan et al., Computational Investigations to Identify Potent Natural Flavonoid Inhibitors of the Nonstructural Protein (NSP) 16/10 Complex Against Coronavirus, Cureus, doi:10.7759/cureus.68098.
16.
Sunita et al., Characterization of Phytochemical Inhibitors of the COVID-19 Primary Protease Using Molecular Modelling Approach, Asian Journal of Microbiology and Biotechnology, doi:10.56557/ajmab/2024/v9i28800.
17.
Wu et al., Biomarkers Prediction and Immune Landscape in Covid-19 and “Brain Fog”, Elsevier BV, doi:10.2139/ssrn.4897774.
18.
Raman et al., Phytoconstituents of Citrus limon (Lemon) as Potential Inhibitors Against Multi Targets of SARS‐CoV‐2 by Use of Molecular Modelling and In Vitro Determination Approaches, ChemistryOpen, doi:10.1002/open.202300198.
19.
Asad et al., Exploring the antiviral activity of Adhatoda beddomei bioactive compounds in interaction with coronavirus spike protein, Archives of Medical Reports, 1:1, archmedrep.com/index.php/amr/article/view/3.
20.
Irfan et al., Phytoconstituents of Artemisia Annua as potential inhibitors of SARS CoV2 main protease: an in silico study, BMC Infectious Diseases, doi:10.1186/s12879-024-09387-w.
21.
Yuan et al., Network pharmacology and molecular docking reveal the mechanisms of action of Panax notoginseng against post-COVID-19 thromboembolism, Review of Clinical Pharmacology and Pharmacokinetics - International Edition, doi:10.61873/DTFA3974.
22.
Nalban et al., Targeting COVID-19 (SARS-CoV-2) main protease through phytochemicals of Albizia lebbeck: molecular docking, molecular dynamics simulation, MM–PBSA free energy calculations, and DFT analysis, Journal of Proteins and Proteomics, doi:10.1007/s42485-024-00136-w.
23.
Zhou et al., Bioinformatics and system biology approaches to determine the connection of SARS-CoV-2 infection and intrahepatic cholangiocarcinoma, PLOS ONE, doi:10.1371/journal.pone.0300441.
24.
Waqas et al., Discovery of Novel Natural Inhibitors Against SARS-CoV-2 Main Protease: A Rational Approach to Antiviral Therapeutics, Current Medicinal Chemistry, doi:10.2174/0109298673292839240329081008.
25.
Hasanah et al., Decoding the therapeutic potential of empon-empon: a bioinformatics expedition unraveling mechanisms against COVID-19 and atherosclerosis, International Journal of Applied Pharmaceutics, doi:10.22159/ijap.2024v16i2.50128.
26.
Shaik et al., Computational identification of selected bioactive compounds from Cedrus deodara as inhibitors against SARS-CoV-2 main protease: a pharmacoinformatics study, Indian Drugs, doi:10.53879/id.61.02.13859.
27.
Wang et al., Investigating the Mechanism of Qu Du Qiang Fei 1 Hao Fang Formula against Coronavirus Disease 2019 Based on Network Pharmacology Method, World Journal of Traditional Chinese Medicine, doi:10.4103/2311-8571.395061.
28.
Singh et al., Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach, Research Square, doi:10.21203/rs.3.rs-3888947/v1.
29.
El-Megharbel et al., Chemical and spectroscopic characterization of (Artemisinin/Quercetin/ Zinc) novel mixed ligand complex with assessment of its potent high antiviral activity against SARS-CoV-2 and antioxidant capacity against toxicity induced by acrylamide in male rats, PeerJ, doi:10.7717/peerj.15638.
30.
Akinwumi et al., Evaluation of therapeutic potentials of some bioactive compounds in selected African plants targeting main protease (Mpro) in SARS-CoV-2: a molecular docking study, Egyptian Journal of Medical Human Genetics, doi:10.1186/s43042-023-00456-4.
31.
Yang et al., Active ingredient and mechanistic analysis of traditional Chinese medicine formulas for the prevention and treatment of COVID-19: Insights from bioinformatics and in vitro experiments, Medicine, doi:10.1097/MD.0000000000036238.
32.
Chandran et al., Molecular docking analysis of quercetin with known CoVid-19 targets, Bioinformation, doi:10.6026/973206300191081.
33.
Qin et al., Exploring the bioactive compounds of Feiduqing formula for the prevention and management of COVID-19 through network pharmacology and molecular docking, Medical Data Mining, doi:10.53388/MDM202407003.
34.
Moschovou et al., Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms242115894.
35.
Pan (B) et al., Quercetin: A promising drug candidate against the potential SARS-CoV-2-Spike mutants with high viral infectivity, Computational and Structural Biotechnology Journal, doi:10.1016/j.csbj.2023.10.029.
36.
Ahmed et al., Evaluation of the Effect of Zinc, Quercetin, Bromelain and Vitamin C on COVID-19 Patients, International Journal of Diabetes Management, doi:10.61797/ijdm.v2i2.259.
37.
Thapa et al., In-silico Approach for Predicting the Inhibitory Effect of Home Remedies on Severe Acute Respiratory Syndrome Coronavirus-2, Makara Journal of Science, doi:10.7454/mss.v27i3.1609.
38.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
39.
Singh (B) et al., Flavonoids as Potent Inhibitor of SARS-CoV-2 Nsp13 Helicase: Grid Based Docking Approach, Middle East Research Journal of Pharmaceutical Sciences, doi:10.36348/merjps.2023.v03i04.001.
40.
Mandal et al., In silico anti-viral assessment of phytoconstituents in a traditional (Siddha Medicine) polyherbal formulation – Targeting Mpro and pan-coronavirus post-fusion Spike protein, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2023.07.004.
41.
Sai Ramesh et al., Computational analysis of the phytocompounds of Mimusops elengi against spike protein of SARS CoV2 – An Insilico model, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2023.125553.
42.
Corbo et al., Inhibitory potential of phytochemicals on five SARS-CoV-2 proteins: in silico evaluation of endemic plants of Bosnia and Herzegovina, Biotechnology & Biotechnological Equipment, doi:10.1080/13102818.2023.2222196.
43.
Azmi et al., Utilization of quercetin flavonoid compounds in onion (Allium cepa L.) as an inhibitor of SARS-CoV-2 spike protein against ACE2 receptors, 11th International Seminar on New Paradigm and Innovation on Natural Sciences and its Application, doi:10.1063/5.0140285.
44.
Alanzi et al., Structure-based virtual identification of natural inhibitors of SARS-CoV-2 and its Delta and Omicron variant proteins, Future Virology, doi:10.2217/fvl-2022-0184.
45.
Yang (B) et al., In silico evidence implicating novel mechanisms of Prunella vulgaris L. as a potential botanical drug against COVID-19-associated acute kidney injury, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1188086.
46.
Wang (B) et al., Computational Analysis of Lianhua Qingwen as an Adjuvant Treatment in Patients with COVID-19, Society of Toxicology Conference, 2023, www.researchgate.net/publication/370491709_Y_Wang_A_E_Tan_O_Chew_A_Hsueh_and_D_E_Johnson_2023_Computational_Analysis_of_Lianhua_Qingwen_as_an_Adjuvant_Treatment_in_Patients_with_COVID-19_Toxicologist_1921_507.
47.
Ibeh et al., Computational studies of potential antiviral compounds from some selected Nigerian medicinal plants against SARS-CoV-2 proteins, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2023.101230.
48.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
49.
Alavi et al., Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study, Biomedicines, doi:10.3390/biomedicines10123074.
50.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
51.
Şimşek et al., In silico identification of SARS-CoV-2 cell entry inhibitors from selected natural antivirals, Journal of Molecular Graphics and Modelling, doi:10.1016/j.jmgm.2021.108038.
52.
Kandeil et al., Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2, Pathogens, doi:10.3390/pathogens10060758.
53.
Rehman et al., Natural Compounds as Inhibitors of SARS-CoV-2 Main Protease (3CLpro): A Molecular Docking and Simulation Approach to Combat COVID-19, Current Pharmaceutical Design, doi:10.2174/1381612826999201116195851.
54.
Sekiou et al., In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2, ChemRxiv, doi:10.26434/chemrxiv.12181404.v1.
55.
Zhang et al., In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus, Journal of Integrative Medicine, doi:10.1016/j.joim.2020.02.005.
56.
Sisti et al., Evaluation of respiratory virus transmissibility and resilience from fomites: the case of 11 SARS-CoV-2 clinical isolates, Applied and Environmental Microbiology, doi:10.1128/aem.00774-25.
57.
Spinelli et al., Amphibian‐Derived Peptides as Natural Inhibitors of SARS‐CoV‐2 Main Protease (Mpro): A Combined In Vitro and In Silico Approach, Chemistry & Biodiversity, doi:10.1002/cbdv.202403202.
58.
Aguilera-Rodriguez et al., Inhibition of SARS-CoV-2 3CLpro by chemically modified tyrosinase from Agaricus bisporus, RSC Medicinal Chemistry, doi:10.1039/D4MD00289J.
59.
Emam et al., Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors, AMB Express, doi:10.1186/s13568-024-01739-8.
60.
Fang et al., Development of nanoparticles incorporated with quercetin and ACE2-membrane as a novel therapy for COVID-19, Journal of Nanobiotechnology, doi:10.1186/s12951-024-02435-2.
61.
Roy et al., Quercetin inhibits SARS-CoV-2 infection and prevents syncytium formation by cells co-expressing the viral spike protein and human ACE2, Virology Journal, doi:10.1186/s12985-024-02299-w.
62.
DiGuilio et al., Quercetin improves and protects Calu-3 airway epithelial barrier function, Frontiers in Cell and Developmental Biology, doi:10.3389/fcell.2023.1271201.
63.
Zhang (B) et al., Discovery of the covalent SARS‐CoV‐2 Mpro inhibitors from antiviral herbs via integrating target‐based high‐throughput screening and chemoproteomic approaches, Journal of Medical Virology, doi:10.1002/jmv.29208.
64.
Wu (B) et al., SARS-CoV-2 N protein induced acute kidney injury in diabetic db/db mice is associated with a Mincle-dependent M1 macrophage activation, Frontiers in Immunology, doi:10.3389/fimmu.2023.1264447.
65.
Xu (B) et al., Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19, Proceedings of the National Academy of Sciences, doi:10.1073/pnas.2301775120.
66.
Fam et al., Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Scientific Reports, doi:10.1038/s41598-023-31764-9.
67.
Aguado et al., Senolytic therapy alleviates physiological human brain aging and COVID-19 neuropathology, bioRxiv, doi:10.1101/2023.01.17.524329.
68.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
69.
Munafò et al., Quercetin and Luteolin Are Single-digit Micromolar Inhibitors of the SARS-CoV-2 RNA-dependent RNA Polymerase, Research Square, doi:10.21203/rs.3.rs-1149846/v1.
70.
Singh (C) et al., The spike protein of SARS-CoV-2 virus induces heme oxygenase-1: Pathophysiologic implications, Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, doi:10.1016/j.bbadis.2021.166322.
71.
Bahun et al., Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols, Food Chemistry, doi:10.1016/j.foodchem.2021.131594.
72.
Abian et al., Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.07.235.
73.
Shaker et al., Anti-cytokine Storm Activity of Fraxin, Quercetin, and their Combination on Lipopolysaccharide-Induced Cytokine Storm in Mice: Implications in COVID-19, Iranian Journal of Medical Sciences, doi:10.30476/ijms.2023.98947.3102.
74.
Wu (C) et al., Treatment with Quercetin inhibits SARS-CoV-2 N protein-induced acute kidney injury by blocking Smad3-dependent G1 cell cycle arrest, Molecular Therapy, doi:10.1016/j.ymthe.2022.12.002.
75.
Azmi (B) et al., The role of vitamin D receptor and IL‐6 in COVID‐19, Molecular Genetics & Genomic Medicine, doi:10.1002/mgg3.2172.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
g.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
h.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
i.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
j.
The endoribonuclease, also known as NendoU or nsp15, cleaves specific sequences in viral RNA which may help the virus evade detection by the host immune system. Inhibition may hinder the virus's ability to mask itself from the immune system, facilitating a stronger immune response.
k.
The NSP16/10 complex consists of non-structural proteins 16 and 10, forming a 2'-O-methyltransferase that modifies the viral RNA cap structure. This modification helps the virus evade host immune detection by mimicking host mRNA, making NSP16/10 a promising antiviral target.
l.
Cathepsin L is a host lysosomal cysteine protease that can prime the spike protein through an alternative pathway when TMPRSS2 is unavailable. Dual targeting of cathepsin L and TMPRSS2 may maximize disruption of alternative pathways for virus entry.
m.
Wingless-related integration site (Wnt) ligand 3 is a host signaling molecule that activates the Wnt signaling pathway, which is important in development, cell growth, and tissue repair. Some studies suggest that SARS-CoV-2 infection may interfere with the Wnt signaling pathway, and that Wnt3a is involved in SARS-CoV-2 entry.
n.
The frizzled (FZD) receptor is a host transmembrane receptor that binds Wnt ligands, initiating the Wnt signaling cascade. FZD serves as a co-receptor, along with ACE2, in some proposed mechanisms of SARS-CoV-2 infection. The virus may take advantage of this pathway as an alternative entry route.
o.
Low-density lipoprotein receptor-related protein 6 is a cell surface co-receptor essential for Wnt signaling. LRP6 acts in tandem with FZD for signal transduction and has been discussed as a potential co-receptor for SARS-CoV-2 entry.
p.
The ezrin protein links the cell membrane to the cytoskeleton (the cell's internal support structure) and plays a role in cell shape, movement, adhesion, and signaling. Drugs that occupy the same spot on ezrin where the viral spike protein would bind may hindering viral attachment, and drug binding could further stabilize ezrin, strengthening its potential natural capacity to impede viral fusion and entry.
q.
The Adipocyte Differentiation-Related Protein (ADRP, also known as Perilipin 2 or PLIN2) is a lipid droplet protein regulating the storage and breakdown of fats in cells. SARS-CoV-2 may hijack the lipid handling machinery of host cells and ADRP may play a role in this process. Disrupting ADRP's interaction with the virus may hinder the virus's ability to use lipids for replication and assembly.
r.
Neuropilin-1 (NRP1) is a cell surface receptor with roles in blood vessel development, nerve cell guidance, and immune responses. NRP1 may function as a co-receptor for SARS-CoV-2, facilitating viral entry into cells. Blocking NRP1 may disrupt an alternative route of viral entry.
s.
EP300 (E1A Binding Protein P300) is a transcriptional coactivator involved in several cellular processes, including growth, differentiation, and apoptosis, through its acetyltransferase activity that modifies histones and non-histone proteins. EP300 facilitates viral entry into cells and upregulates inflammatory cytokine production.
t.
Prostaglandin G/H synthase 2 (PTGS2, also known as COX-2) is an enzyme crucial for the production of inflammatory molecules called prostaglandins. PTGS2 plays a role in the inflammatory response that can become severe in COVID-19 and inhibitors (like some NSAIDs) may have benefits in dampening harmful inflammation, but note that prostaglandins have diverse physiological functions.
u.
Heat Shock Protein 90 Alpha Family Class A Member 1 (HSP90AA1) is a chaperone protein that helps other proteins fold correctly and maintains their stability. HSP90AA1 plays roles in cell signaling, survival, and immune responses. HSP90AA1 may interact with numerous viral proteins, but note that it has diverse physiological functions.
v.
Matrix metalloproteinase 9 (MMP9), also called gelatinase B, is a zinc-dependent enzyme that breaks down collagen and other components of the extracellular matrix. MMP9 levels increase in severe COVID-19. Overactive MMP9 can damage lung tissue and worsen inflammation. Inhibition of MMP9 may prevent excessive tissue damage and help regulate the inflammatory response.
w.
The interleukin-6 (IL-6) pro-inflammatory cytokine (signaling molecule) has a complex role in the immune response and may trigger and perpetuate inflammation. Elevated IL-6 levels are associated with severe COVID-19 cases and cytokine storm. Anti-IL-6 therapies may be beneficial in reducing excessive inflammation in severe COVID-19 cases.
x.
The interleukin-10 (IL-10) anti-inflammatory cytokine helps regulate and dampen immune responses, preventing excessive inflammation. IL-10 levels can also be elevated in severe COVID-19. IL-10 could either help control harmful inflammation or potentially contribute to immune suppression.
y.
Vascular Endothelial Growth Factor A (VEGFA) promotes the growth of new blood vessels (angiogenesis) and has roles in inflammation and immune responses. VEGFA may contribute to blood vessel leakiness and excessive inflammation associated with severe COVID-19.
z.
RELA is a transcription factor subunit of NF-kB and is a key regulator of inflammation, driving pro-inflammatory gene expression. SARS-CoV-2 may hijack and modulate NF-kB pathways.
aa.
The interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor is a primary method of viral entry, inhibiting this interaction can prevent the virus from attaching to and entering host cells, halting infection at an early stage.
ab.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
ac.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
ad.
HEK293-ACE2+ is a human embryonic kidney cell line engineered for high ACE2 expression and SARS-CoV-2 susceptibility.
ae.
Huh-7 cells were derived from a liver tumor (hepatoma).
af.
Caco-2 cells come from a colorectal adenocarcinoma (cancer). They are valued for their ability to form a polarized cell layer with properties similar to the intestinal lining.
ag.
Vero E6 is an African green monkey kidney cell line with low/no ACE2 expression and high SARS-CoV-2 susceptibility. The cell line is easy to maintain and supports robust viral replication, however the monkey origin may not accurately represent human responses.
ah.
mTEC is a mouse tubular epithelial cell line.
ai.
RAW264.7 is a mouse macrophage cell line.
aj.
HLMEC (Human Lung Microvascular Endothelial Cells) are primary endothelial cells derived from the lung microvasculature. They are used to study endothelial function, inflammation, and viral interactions, particularly in the context of lung infections such as SARS-CoV-2. HLMEC express ACE2 and are susceptible to SARS-CoV-2 infection, making them a relevant model for studying viral entry and endothelial responses in the lung.
ak.
A mouse model expressing the human ACE2 receptor under the control of the K18 promoter.
al.
A mouse model of obesity and severe insulin resistance leading to type 2 diabetes due to a mutation in the leptin receptor gene that impairs satiety signaling.
am.
A mouse model commonly used in infectious disease and cancer research due to higher immune response and susceptibility to infection.
Shaik et al., 28 Feb 2024, peer-reviewed, 3 authors.
Contact: aminaammi786@gmail.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
COMPUTATIONAL IDENTIFICATION OF SELECTED BIOACTIVE COMPOUNDS FROM CEDRUS DEODARA AS INHIBITORS AGAINST SARS-COV-2 MAIN PROTEASE: A PHARMACOINFORMATICS STUDY
INDIAN DRUGS, doi:10.53879/id.61.02.13859
Amid the ongoing Covid-19 pandemic, the quest for potent antiviral treatments intensifies. This study focuses on the potential of bioactive compounds from the Himalayan cedar Cedrus deodara against the SARS-CoV-2 virus. Specifically targeting the main protease (M Pro ) and spike protein, the study employs docking trials and molecular dynamics simulations. Compounds such as quercetin, dihydrodehydrodiconiferyl alcohol, and cedeodarin exhibit notable binding affinity, surpassing the reference drug favipiravir. Molecular dynamics simulations affirm the stability of these complexes throughout the simulation period. While these findings underscore promising interactions, it is crucial to emphasize the need for further research and experimental validation to fully explore the therapeutic capabilities of C. deodara in combatting Covid-19.
References
Aminabee, Raveesha, Adithya, Mohansai, Shaherbanu et al., In vivo antinociceptive activity and In silico molecular docking of selected phytoconstituents of methanolic extract of Hypericum japonicum, J. Drug Alcohol Res
Available, Idma, None
Chandramohan, Kaphle, Chekuri, Evaluating andrographolide as a potent inhibitor of NS3-4A protease and its drug resistant mutants using in silico approaches, Adv. Virol
Chaudhuri, Symons, Deval, Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond, Antivir. Res
Cheng, Li, Zhou, AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties, J. Chem. Inf. Model
Du, Xu, Structure of Mpro from COVID-19 virus and discovery of its inhibitors
Gangadharappa, Sharath, Revanasiddappa, Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors, J. Biomol. Struct. Dyn
Goel, Singh, Lagunin, PASS-assisted exploration of new therapeutic potential of natural products, Med. Chem. Res
Hui, Azhar, Madani, The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health -The latest 2019 novel coronavirus outbreak in Wuhan, China, Int. J. Infect. Dis
Hussain, Haleem, Khan, Medicinal plants: a repository of antiviral metabolites, Future Virol
Jassim, Naji, Novel antiviral agents: a medicinal plant perspective, J. Appl. Microbiol
Kar, Sharma, Singh, Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: An in silico investigation, J Biomol. Struct. Dyn
Khurana, Ishar, Gajbhiye, PASS assisted prediction and pharmacological evaluation of novel nicotinic analogs for nootropic activity in mice, Eur. J. Pharmacol
Lin, Hsu, Lin, Antiviral natural products and herbal medicines, J. Tradit. Complem. Med
Lu, Zhao, Li, Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet
Martinez, Sasse, Bronstrup, Antiviral drug discovery: broad-spectrum drugs from nature, Nat. Prod. Rep
Mittal, Goel, Bhargava, PASS-assisted exploration of antidepressant activity of 1,3,4-trisubstituted-β-lactam derivatives, Bioorg. Med. Chem. Lett
Morris, Huey, Lindstrom, Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem
Moses, Goossens, Plants for human health: greening biotechnology and synthetic biology, J. Exp. Bot
O'boyle, Banck, James, Open Babel: an open chemical toolbox, J Cheminform
Pangastuti, Amin, Amin, Natural bioactive compound from Moringa oleifera against cancer based on in silico screening, J. Teknol
Prasanth, Aminabee, Rao, Guntupalli Ch, Reddy et al., Inhibitory effects of Manosa alliacea in Freund's adjuvant arthritis on inflammatory markers and its confirmation by In silico strategy, Thai J. Pharm. Sci
Prasanth, Aminabee, Rao, Teja, Bhargavi et al., Antihelmintic activity of Mansoa alliacea against Pheretima posthuma: In vitro and In silico approach, Thai J. Pharm. Sci
Rouchka, Chariker, Chung, Phylogenetic and variant analysis of 1,040 SARS-CoV-2 Genomes
Sander, Freyss, Korff, Datawarrior: an open-source program for chemistry aware data visualization and analysis, J. Chem. Inf. Model
Schaal, Plants and people: our shared history and future, Plants People Planet
Seeliger, Groot, Ligand docking and binding site analysis with PyMOL and Autodock/Vina, J. Comput. Aided Mol. Des
Shaik, Atmakuri, Clinical trials status and approaches of COVID-19 vaccines developed globally: The Recent Updates, Pharma Times
Shaik, Atmakuri, Maram, In vivo antioxidant activity of different fractions of Indigofera barberi against paracetamol induced toxicity in rats, Turk. J. Pharm. Sci
Sussman, Lin, Jiang, Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules, Acta Crystallogr. D Biol. Crystallogr
To, Hung, Chan, From SARS coronavirus to novel animal and human coronaviruses, J. Thorac. Dis
Van Der Spoel, Lindahl, Hess, GROMACS: fast, flexible, and free, J. Comput. Chem
Yang, Lou, Sun, AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties, Bioinformatics
Zhou, Hou, Shen, Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2, Cell Discov
Zhu, Zhang, Wang, A novel coronavirus from patients with pneumonia in China, New Engl. J. Med
DOI record:
{
"DOI": "10.53879/id.61.02.13859",
"ISSN": [
"0019-462X"
],
"URL": "http://dx.doi.org/10.53879/id.61.02.13859",
"abstract": "<jats:p>Amid the ongoing Covid-19 pandemic, the quest for potent antiviral treatments intensifies. This study focuses on the potential of bioactive compounds from the Himalayan cedar Cedrus deodara against the SARS-CoV-2 virus. Specifically targeting the main protease (MPro) and spike protein, the study employs docking trials and molecular dynamics simulations. Compounds such as quercetin, dihydrodehydrodiconiferyl alcohol, and cedeodarin exhibit notable binding affinity, surpassing the reference drug favipiravir. Molecular dynamics simulations affirm the stability of these complexes throughout the simulation period. While these findings underscore promising interactions, it is crucial to emphasize the need for further research and experimental validation to fully explore the therapeutic capabilities of C. deodara in combatting Covid-19.</jats:p>",
"author": [
{
"affiliation": [],
"family": "Shaik",
"given": "Aminabee",
"sequence": "first"
},
{
"affiliation": [],
"name": "Department of Pharmacology, V. V. Institute of Pharmaceutical Sciences, Gudlavalleru-521 356, Andhra Pradesh, India",
"sequence": "first"
},
{
"affiliation": [],
"family": "Atmakuri",
"given": "Lakshmana Rao",
"sequence": "additional"
}
],
"container-title": "INDIAN DRUGS",
"container-title-short": "IND. DRU.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
3,
18
]
],
"date-time": "2024-03-18T13:23:54Z",
"timestamp": 1710768234000
},
"deposited": {
"date-parts": [
[
2024,
3,
20
]
],
"date-time": "2024-03-20T10:59:47Z",
"timestamp": 1710932387000
},
"indexed": {
"date-parts": [
[
2024,
3,
21
]
],
"date-time": "2024-03-21T01:28:16Z",
"timestamp": 1710984496790
},
"is-referenced-by-count": 0,
"issue": "02",
"issued": {
"date-parts": [
[
2024,
2,
28
]
]
},
"journal-issue": {
"issue": "02",
"published-online": {
"date-parts": [
[
2024,
3,
14
]
]
},
"published-print": {
"date-parts": [
[
2024,
2,
28
]
]
}
},
"link": [
{
"URL": "https://www.indiandrugsonline.org//download/February_2024_61_2_78-91.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "31700",
"original-title": [],
"page": "78-91",
"prefix": "10.53879",
"published": {
"date-parts": [
[
2024,
2,
28
]
]
},
"published-online": {
"date-parts": [
[
2024,
3,
14
]
]
},
"published-print": {
"date-parts": [
[
2024,
2,
28
]
]
},
"publisher": "Indian Drug Manufacturers' Association (IDMA)",
"reference": [
{
"key": "ref0",
"unstructured": "1. World Health Organization. Coronavirus disease 2019 (COVID19): Situation report. 2020, 70."
},
{
"DOI": "10.13005/bpj/2214",
"doi-asserted-by": "publisher",
"key": "ref1",
"unstructured": "2. Shaik A., and Atmakuri L.R.: Clinical trials status and approaches of COVID-19 vaccines developed globally: The Recent Updates. Pharma Times, 2022, 54(3), 7-14."
},
{
"DOI": "10.20944/preprints202005.0396.v1",
"doi-asserted-by": "publisher",
"key": "ref2",
"unstructured": "3. Rouchka E.C., Chariker J.H. and Chung D.: Phylogenetic and variant analysis of 1,040 SARS-CoV-2 Genomes. 2020"
},
{
"key": "ref3",
"unstructured": "4. Lu R., Zhao X. and Li J.: Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet, 2020, 395, 565-574."
},
{
"key": "ref4",
"unstructured": "5. Jin Z., Du X. and Xu Y.: Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv. Preprint. 2020"
},
{
"DOI": "10.1016/j.ijid.2020.01.009",
"doi-asserted-by": "publisher",
"key": "ref5",
"unstructured": "6. Hui D.S., Azhar E.I. and Madani T.A.: The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis., 2020, 91, 264-266."
},
{
"DOI": "10.1093/jxb/erx268",
"doi-asserted-by": "publisher",
"key": "ref6",
"unstructured": "7. Moses T. and Goossens A.: Plants for human health: greening biotechnology and synthetic biology. J. Exp. Bot., 2017, 68, 4009-4011."
},
{
"DOI": "10.1002/ppp3.12",
"doi-asserted-by": "publisher",
"key": "ref7",
"unstructured": "8. Schaal B.: Plants and people: our shared history and future. Plants People Planet, 2019, 1, 14-19."
},
{
"DOI": "10.1046/j.1365-2672.2003.02026.x",
"doi-asserted-by": "publisher",
"key": "ref8",
"unstructured": "9. Jassim S.A.A. and Naji M. A.: Novel antiviral agents: a medicinal plant perspective. J. Appl. Microbiol., 2003, 95, 412-427."
},
{
"DOI": "10.2217/fvl-2016-0110",
"doi-asserted-by": "publisher",
"key": "ref9",
"unstructured": "10. Hussain W., Haleem K.S. and Khan I.: Medicinal plants: a repository of antiviral metabolites. Future Virol., 2017, 12, 299-308."
},
{
"DOI": "10.4274/tjps.galenos.2018.30306",
"doi-asserted-by": "publisher",
"key": "ref10",
"unstructured": "11. Shaik A., Atmakuri L.R. and Maram C.E.: In vivo antioxidant activity of different fractions of Indigofera barberi against paracetamol induced toxicity in rats. Turk. J. Pharm. Sci., 2020, 17(2), 136-140."
},
{
"DOI": "10.1016/j.antiviral.2018.05.005",
"doi-asserted-by": "publisher",
"key": "ref11",
"unstructured": "12. Chaudhuri S., Symons J.A. and Deval J.: Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond. Antivir. Res., 2018, 155, 76-88."
},
{
"DOI": "10.1021/ci500588j",
"doi-asserted-by": "publisher",
"key": "ref12",
"unstructured": "13. Sander T., Freyss J. and Von Korff M.: Datawarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model, 2015, 55, 460-473."
},
{
"DOI": "10.1107/S0907444998009378",
"doi-asserted-by": "publisher",
"key": "ref13",
"unstructured": "14. Sussman J.L., Lin D. and Jiang J.: Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr., 1998, 54, 1078-1084."
},
{
"key": "ref14",
"unstructured": "15. Aminabee S.K., Raveesha P., Adithya V., Mohansai M., Shaherbanu, Harshitha K., Himaja Kasthuri K., Lakshmi Priya M., Chandini Naga Mallika G. and Lakshmana Rao A.: In vivo antinociceptive activity and In silico molecular docking of selected phytoconstituents of methanolic extract of Hypericum japonicum. J. Drug Alcohol Res., 2022, 11(5), 01-08."
},
{
"DOI": "10.1080/07391102.2020.1780947",
"doi-asserted-by": "publisher",
"key": "ref15",
"unstructured": "16. Kar P., Sharma N.R. and Singh B.: Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: An in silico investigation. J Biomol. Struct. Dyn., 2020, 1-12."
},
{
"DOI": "10.1002/jcc.21256",
"doi-asserted-by": "publisher",
"key": "ref16",
"unstructured": "17. Morris G.M., Huey R. and Lindstrom W.: Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem., 2009, 30, 2785-2791."
},
{
"DOI": "10.1186/1758-2946-3-33",
"doi-asserted-by": "publisher",
"key": "ref17",
"unstructured": "18. O'Boyle N.M., Banck M. and James C.A. Open Babel: an open chemical toolbox. J Cheminform., 2011, 3, 33."
},
{
"DOI": "10.11113/jt.v78.8328",
"doi-asserted-by": "publisher",
"key": "ref18",
"unstructured": "19. Pangastuti A., Amin I. F. and Amin A.Z.: Natural bioactive compound from Moringa oleifera against cancer based on in silico screening. J. Teknol., 2016, 78, 315-318."
},
{
"key": "ref19",
"unstructured": "20. Prasanth D.S.N.B.K., Aminabee S.K., Lakshmana Rao A., Guntupalli Ch., Rajasekhar Reddy A., Umasankar K., Koteswara Rao S.N., Rajeshwari P.: Inhibitory effects of Manosa alliacea in Freund's adjuvant arthritis on inflammatory markers and its confirmation by In silico strategy. Thai J. Pharm. Sci., 2021, 45(6), 532-544."
},
{
"DOI": "10.1007/s10822-010-9352-6",
"doi-asserted-by": "publisher",
"key": "ref20",
"unstructured": "21. Seeliger D. and De Groot B.L.: Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des., 2010, 24, 417-422."
},
{
"DOI": "10.1093/bioinformatics/bty707",
"doi-asserted-by": "publisher",
"key": "ref21",
"unstructured": "22. Yang H., Lou C. and Sun L.: AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics., 2019, 35, 1067-1069."
},
{
"DOI": "10.1021/ci300367a",
"doi-asserted-by": "publisher",
"key": "ref22",
"unstructured": "23. Cheng F., Li W. and Zhou Y.: AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model., 2012, 52, 3099-3105."
},
{
"DOI": "10.1016/j.ejphar.2011.04.048",
"doi-asserted-by": "publisher",
"key": "ref23",
"unstructured": "24. Khurana N., Ishar M.P.S. and Gajbhiye A.: PASS assisted prediction and pharmacological evaluation of novel nicotinic analogs for nootropic activity in mice. Eur. J. Pharmacol., 2011, 662, 22-30."
},
{
"DOI": "10.1016/j.bmcl.2008.09.064",
"doi-asserted-by": "publisher",
"key": "ref24",
"unstructured": "25. Mittal M., Goel R.K. and Bhargava G.: PASS-assisted exploration of antidepressant activity of 1,3,4-trisubstituted-β-lactam derivatives. Bioorg. Med. Chem. Lett., 2008, 18, 5347-5349."
},
{
"DOI": "10.1007/s00044-010-9398-y",
"doi-asserted-by": "publisher",
"key": "ref25",
"unstructured": "26. Goel R.K., Singh D. and Lagunin A.: PASS-assisted exploration of new therapeutic potential of natural products. Med. Chem. Res., 2011, 20, 1509-1514."
},
{
"DOI": "10.1002/jcc.20291",
"doi-asserted-by": "publisher",
"key": "ref26",
"unstructured": "27. Van Der Spoel D., Lindahl E. and Hess B.: GROMACS: fast, flexible, and free. J. Comput. Chem., 2005, 26, 1701-1718."
},
{
"DOI": "10.1080/07391102.2019.1667265",
"doi-asserted-by": "publisher",
"key": "ref27",
"unstructured": "28. Gangadharappa B.S., Sharath R. and Revanasiddappa P.D.: Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors. J. Biomol. Struct. Dyn., 2020, 38, 3757-3771."
},
{
"DOI": "10.1155/2015/972067",
"doi-asserted-by": "publisher",
"key": "ref28",
"unstructured": "29. Chandramohan V., Kaphle A. and Chekuri M.: Evaluating andrographolide as a potent inhibitor of NS3-4A protease and its drug resistant mutants using in silico approaches. Adv. Virol., 2015, 15, 20-67."
},
{
"key": "ref29",
"unstructured": "30. Prasanth D.S.N.B.K., Aminabee S.K., Lakshmana Rao A., Teja N., Bhargavi K., Monika Ch., Pujitha B., Sandhya T., Lalitha A. and Siva Prasad P.: Antihelmintic activity of Mansoa alliacea against Pheretima posthuma: In vitro and In silico approach. Thai J. Pharm. Sci., 2020, 44(3), 86-196."
},
{
"key": "ref30",
"unstructured": "31. To K.K., Hung I.F. and Chan J.F.: From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis., 2013, 5(2), S103-S108."
},
{
"DOI": "10.1056/NEJMoa2001017",
"doi-asserted-by": "publisher",
"key": "ref31",
"unstructured": "32. Zhu N., Zhang D. and Wang W.: A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med., 2020, 82, 727-733."
},
{
"DOI": "10.1038/s41421-020-0153-3",
"doi-asserted-by": "publisher",
"key": "ref32",
"unstructured": "33. Zhou Y., Hou Y. and Shen J.: Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov., 2020, 6, 14."
},
{
"DOI": "10.4103/2225-4110.124335",
"doi-asserted-by": "publisher",
"key": "ref33",
"unstructured": "34. Lin L.T., Hsu W.C. and Lin C.C.: Antiviral natural products and herbal medicines. J. Tradit. Complement. Med., 2014, 4, 24-35."
},
{
"DOI": "10.1039/C4NP00085D",
"doi-asserted-by": "publisher",
"key": "ref34",
"unstructured": "35. Martinez J.P., Sasse F. and Bronstrup M.: Antiviral drug discovery: broad-spectrum drugs from nature. Nat. Prod. Rep., 2015, 32, 29-48."
}
],
"reference-count": 35,
"references-count": 35,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.indiandrugsonline.org/issuesarticle-details?id=MTU0Nw=="
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Drug Discovery",
"Pharmaceutical Science",
"Pharmacology"
],
"subtitle": [],
"title": "COMPUTATIONAL IDENTIFICATION OF SELECTED BIOACTIVE COMPOUNDS FROM CEDRUS DEODARA AS INHIBITORS AGAINST SARS-COV-2 MAIN PROTEASE: A PHARMACOINFORMATICS STUDY",
"type": "journal-article",
"volume": "61"
}