Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2
et al., Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908, Jan 2021
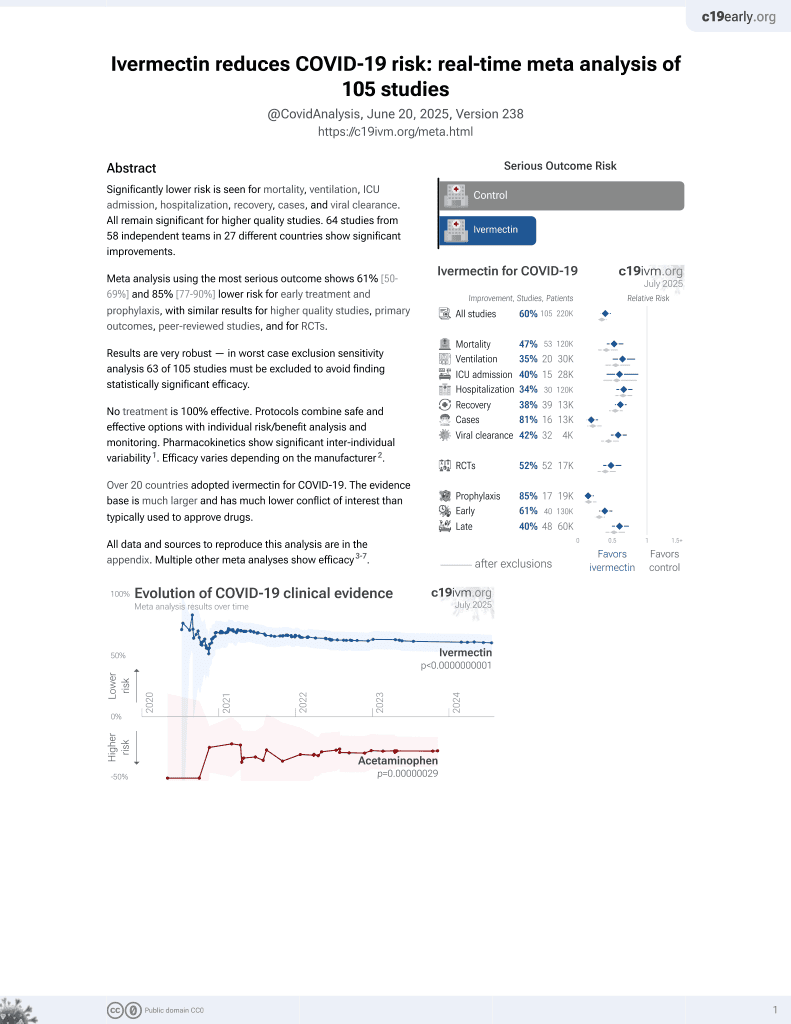
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Molecular docking analysis showing that ivermectin efficiently binds to the viral S protein as well as the human cell surface receptors ACE-2 and TMPRSS2; therefore, it might be involved in inhibiting the entry of the virus into the host cell. It also binds to Mpro and PLpro of SARS-CoV-2; therefore, it might play a role in preventing the post-translational processing of viral polyproteins. The highly efficient binding of ivermectin to the viral N phosphoprotein and nsp14 is suggestive of its role in inhibiting viral replication and assembly.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Eweas et al., 25 Jan 2021, peer-reviewed, 3 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2
Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908
SARS-CoV-2 is a newly emerged coronavirus that causes a respiratory disease with variable severity and fatal consequences. It was first reported in Wuhan and subsequently caused a global pandemic. The viral spike protein binds with the ACE-2 cell surface receptor for entry, while TMPRSS2 triggers its membrane fusion. In addition, RNA dependent RNA polymerase (RdRp), 3 -5 exoribonuclease (nsp14), viral proteases, N, and M proteins are important in different stages of viral replication. Accordingly, they are attractive targets for different antiviral therapeutic agents. Although many antiviral agents have been used in different clinical trials and included in different treatment protocols, the mode of action against SARS-CoV-2 is still not fully understood. Different potential repurposed drugs, including, chloroquine, hydroxychloroquine, ivermectin, remdesivir, and favipiravir, were screened in the present study. Molecular docking of these drugs with different SARS-CoV-2 target proteins, including spike and membrane proteins, RdRp, nucleoproteins, viral proteases, and nsp14, was performed. Moreover, the binding affinities of the human ACE-2 receptor and TMPRSS2 to the different drugs were evaluated. Molecular dynamics simulation and MM-PBSA calculation were also conducted. Ivermectin and remdesivir were found to be the most promising drugs. Our results suggest that both these drugs utilize different mechanisms at the entry and post-entry stages and could be considered potential inhibitors of SARS-CoV-2 replication.
AUTHOR CONTRIBUTIONS ASA-M contributed conception of the study and critically revised the manuscript. AFE conducted the molecular docking and wrote the first draft of the manuscript. AAA performed the molecular dynamics simulation. ASA-M, AFE, and AAA shared analyzing and discussing the results. All authors contributed to manuscript revision, read, and approved the submitted version.
SUPPLEMENTARY MATERIAL The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020. 592908/full#supplementary-material Supplementary Figure 3 | RMSD and number of Hydrogen bonds. For every receptor, the RMSD and Hydrogen bonds number for each ligand were demonstrated.
Conflict of Interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdel-Moneim, Abdelwhab, Evidence for SARS-CoV-2 infection of animal hosts, Pathogens, doi:10.3390/pathogens9070529
Abraham, Murtola, Schulz, Páll, Smith et al., GROMACS: high performance molecular simulations through multilevel parallelism from laptops to supercomputers, SoftwareX, doi:10.1016/j.softx.2015.06.001
Baker, Sept, Joseph, Holst, Mccammon, Electrostatics of nanosystems: application to microtubules and the ribosome, Proc. Natl. Acad. Sci. U.S.A, doi:10.1073/pnas.181342398
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the treatment of Covid-19-preliminary report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Cai, Yang, Liu, Chen, Shu et al., Experimental treatment with favipiravir for COVID-19: an open-label control study, Engineering, doi:10.1016/j.eng.2020.03.007
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res, doi:10.1016/j.antiviral.2020.104787
Choy, Wong, Kaewpreedee, Sia, Chen et al., Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro, Antiviral Res, doi:10.1016/j.antiviral.2020.104786
Deshpande, Tiwari, Nyayanit, Modak, In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2, Eur. J. Pharmacol, doi:10.1016/j.ejphar.2020.173430
Eckerle, Becker, Halpin, Li, Venter et al., Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing, PLoS Pathog, doi:10.1371/journal.ppat.1000896
Fehr, Perlman, Coronaviruses: an overview of their replication and pathogenesis, Methods Mol. Biol, doi:10.1007/978-1-4939-2438-7_1
Gao, Yan, Huang, Liu, Zhao et al., Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science, doi:10.1126/science.abb7498
Glowacka, Bertram, Müller, Allen, Soilleux et al., Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response, J. Virol, doi:10.1128/JVI.02232-10
Gurung, In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors, Gene Rep, doi:10.1016/j.genrep.2020.100860
Hall, Ji, A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease, Travel Med. Infect. Dis, doi:10.1016/j.tmaid.2020.101646
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Hosein, Moore, Known SARS-CoV-2 infections: the tip of an important iceberg, Int. J. Health Plann. Manage, doi:10.1002/hpm.3006
Ivanov, Thiel, Dobbe, Van Der Meer, Snijder et al., Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase, J. Virol, doi:10.1128/JVI.78.11.5619-5632.2004
Kang, Yang, Hong, Zhang, Huang et al., Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites, Acta Pharm. Sin. B, doi:10.1016/j.apsb.2020.04.009
Kumar, Singh, Patel, In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing, J. Infect. Public Health, doi:10.1016/j.jiph.2020.06.016
Kumari, Kumar, g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations, J. Chem. Inf. Model, doi:10.1021/ci500020m
Kwiek, Haystead, Rudolph, Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines, Biochemistry, doi:10.1021/bi035923w
Lehrer, Rheinstein, Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2, Vivo, doi:10.21873/invivo.12134
Ma, Wu, Shaw, Gao, Wang et al., Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex, Proc. Natl. Acad. Sci. U.S.A, doi:10.1073/pnas.1508686112
Maier, Martinez, Kasavajhala, Wickstrom, Hauser et al., ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB, J. Chem. Theory Comput, doi:10.1021/acs.jctc.5b00255
Mielech, Chen, Mesecar, Baker, Nidovirus papainlike proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities, Virus Res, doi:10.1016/j.virusres.2014.01.025
Mugisha, Vuong, Puray-Chavez, Kutluay, A facile Q-RT-PCR assay for monitoring SARS-CoV-2 growth in cell culture, bioRxiv, doi:10.1101/2020.06.26.174698
Osipiuk, Jedrzejczak, Tesar, Endres, Stols et al., The crystal structure of papain-like protease of SARS CoV-2, Center for Structural Genomics of Infectious Diseases, doi:10.2210/pdb6w9c/pdb
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF chimera-a visualization system for exploratory research and analysis, J. Comput. Chem, doi:10.1002/jcc.20084
Rowland, Chauhan, Fang, Pekosz, Kerrigan et al., Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells, J. Virol, doi:10.1128/JVI.79.17.11507-11512.2005
Sali, Blundell, Comparative protein modelling by satisfaction of spatial restraints, J. Mol. Biol, doi:10.1006/jmbi.1993.1626
Savarino, Di Trani, Donatelli, Cauda, Cassone, New insights into the antiviral effects of chloroquine, Lancet Infect. Dis, doi:10.1016/S1473-3099(06)70361-9
Shannon, Selisko, Le, Huchting, Touret et al., Favipiravir strikes the SARS-CoV-2 at its achilles heel, the RNA polymerase, doi:10.1101/2020.05.15.098731
Sheahan, Sims, Leist, Schäfer, Won et al., Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV, Nat. Commun, doi:10.1038/s41467-019-13940-6
Siu, Teoh, Lo, Chan, Kien et al., The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles, J. Virol, doi:10.1128/JVI.01052-08
Sousa Da Silva, Vranken, ACPYPE -AnteChamber PYthon Parser interfacE, BMC Res. Notes, doi:10.1186/1756-0500-5-367
Tay, Fraser, Chan, Moreland, Rathore et al., Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Res, doi:10.1016/j.antiviral.2013.06.002
Timani, Liao, Ye, Zeng, Liu et al., Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus, Virus Res, doi:10.1016/j.virusres.2005.05.007
Vincent, Bergeron, Benjannet, Erickson, Rollin et al., Chloroquine is a potent inhibitor of SARS coronavirus infection and spread, Virol. J, doi:10.1186/1743-422X-2-69
Wagstaff, Sivakumaran, Heaton, Harrich, Jans, Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochem. J, doi:10.1042/BJ20120150
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, doi:10.1016/j.cell.2020.11.032
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res, doi:10.1038/s41422-020-0282-0
Wang, Wang, Kollman, Case, Automatic atom type and bond type perception in molecular mechanical calculations, J. Mol. Graph. Model, doi:10.1016/j.jmgm.2005.12.005
Wang, Zhang, Du, Du, Zhao et al., Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial, Lancet
Warren, Jordan, Lo, Ray, Mackman et al., Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys, Nature
Wu, Liu, Yang, Zhang, Zhong et al., Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharm. Sin. B
Wu, Peng, Huang, Ding, Wang et al., Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China, Cell Host Microbe
Xu, Liu, Weiss, Arnold, Sarafianos et al., Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design, Nucleic Acids Res
Yan, Zou, Sun, Li, Xu et al., Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model, Cell Res
Yang, Atkinson, Wang, Lee, Bogoyevitch et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Res, doi:10.1016/j.antiviral.2020.104760
Yin, Mao, Luan, Shen, Shen et al., Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science, doi:10.1126/science.abc1560
Zhang, Lin, Sun, Curth, Drosten et al., Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, Science, doi:10.1126/science.abb3405
Zhao, Jha, Wu, Elliott, Ziebuhr et al., Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology, Cell Host Microbe, doi:10.1016/j.chom.2012.04.011
Zhu, Zhang, Wang, Li, Yang et al., A novel coronavirus from patients with pneumonia in China, N. Engl. J. Med, doi:10.1056/NEJMoa2001017
Ziebuhr, Snijder, Gorbalenya, Virus-encoded proteinases and proteolytic processing in the Nidovirales, J. Gen. Virol, doi:10.1099/0022-1317-81-4-853
DOI record:
{
"DOI": "10.3389/fmicb.2020.592908",
"ISSN": [
"1664-302X"
],
"URL": "http://dx.doi.org/10.3389/fmicb.2020.592908",
"abstract": "<jats:p>SARS-CoV-2 is a newly emerged coronavirus that causes a respiratory disease with variable severity and fatal consequences. It was first reported in Wuhan and subsequently caused a global pandemic. The viral spike protein binds with the ACE-2 cell surface receptor for entry, while TMPRSS2 triggers its membrane fusion. In addition, RNA dependent RNA polymerase (RdRp), 3′–5′ exoribonuclease (nsp14), viral proteases, N, and M proteins are important in different stages of viral replication. Accordingly, they are attractive targets for different antiviral therapeutic agents. Although many antiviral agents have been used in different clinical trials and included in different treatment protocols, the mode of action against SARS-CoV-2 is still not fully understood. Different potential repurposed drugs, including, chloroquine, hydroxychloroquine, ivermectin, remdesivir, and favipiravir, were screened in the present study. Molecular docking of these drugs with different SARS-CoV-2 target proteins, including spike and membrane proteins, RdRp, nucleoproteins, viral proteases, and nsp14, was performed. Moreover, the binding affinities of the human ACE-2 receptor and TMPRSS2 to the different drugs were evaluated. Molecular dynamics simulation and MM-PBSA calculation were also conducted. Ivermectin and remdesivir were found to be the most promising drugs. Our results suggest that both these drugs utilize different mechanisms at the entry and post-entry stages and could be considered potential inhibitors of SARS-CoV-2 replication.</jats:p>",
"alternative-id": [
"10.3389/fmicb.2020.592908"
],
"author": [
{
"affiliation": [],
"family": "Eweas",
"given": "Ahmad F.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Alhossary",
"given": "Amr A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Abdel-Moneim",
"given": "Ahmed S.",
"sequence": "additional"
}
],
"container-title": "Frontiers in Microbiology",
"container-title-short": "Front. Microbiol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2021,
1,
25
]
],
"date-time": "2021-01-25T05:25:07Z",
"timestamp": 1611552307000
},
"deposited": {
"date-parts": [
[
2021,
2,
22
]
],
"date-time": "2021-02-22T17:09:48Z",
"timestamp": 1614013788000
},
"funder": [
{
"DOI": "10.13039/501100006261",
"doi-asserted-by": "publisher",
"name": "Taif University"
}
],
"indexed": {
"date-parts": [
[
2024,
4,
2
]
],
"date-time": "2024-04-02T10:54:47Z",
"timestamp": 1712055287979
},
"is-referenced-by-count": 62,
"issued": {
"date-parts": [
[
2021,
1,
25
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
1,
25
]
],
"date-time": "2021-01-25T00:00:00Z",
"timestamp": 1611532800000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fmicb.2020.592908/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2021,
1,
25
]
]
},
"published-online": {
"date-parts": [
[
2021,
1,
25
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.3390/pathogens9070529",
"article-title": "Evidence for SARS-CoV-2 infection of animal hosts.",
"author": "Abdel-Moneim",
"doi-asserted-by": "publisher",
"journal-title": "Pathogens",
"key": "B1",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1016/j.softx.2015.06.001",
"article-title": "GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers.",
"author": "Abraham",
"doi-asserted-by": "publisher",
"first-page": "19",
"journal-title": "SoftwareX",
"key": "B2",
"volume": "1",
"year": "2015"
},
{
"DOI": "10.1073/pnas.181342398",
"article-title": "Electrostatics of nanosystems: application to microtubules and the ribosome.",
"author": "Baker",
"doi-asserted-by": "publisher",
"first-page": "10037",
"journal-title": "Proc. Natl. Acad. Sci. U.S.A.",
"key": "B3",
"volume": "98",
"year": "2001"
},
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the treatment of Covid-19—preliminary report.",
"author": "Beigel",
"doi-asserted-by": "publisher",
"first-page": "1813",
"journal-title": "N. Engl. J. Med.",
"key": "B4",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1016/j.eng.2020.03.007",
"article-title": "Experimental treatment with favipiravir for COVID-19: an open-label control study.",
"author": "Cai",
"doi-asserted-by": "publisher",
"first-page": "1192",
"journal-title": "Engineering",
"key": "B5",
"volume": "6",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"article-title": "The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro.",
"author": "Caly",
"doi-asserted-by": "publisher",
"journal-title": "Antiviral Res.",
"key": "B6",
"volume": "178",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2020.104786",
"article-title": "Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro.",
"author": "Choy",
"doi-asserted-by": "publisher",
"journal-title": "Antiviral Res.",
"key": "B7",
"volume": "178",
"year": "2020"
},
{
"DOI": "10.1016/j.ejphar.2020.173430",
"article-title": "In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2.",
"author": "Deshpande",
"doi-asserted-by": "publisher",
"journal-title": "Eur. J. Pharmacol.",
"key": "B8",
"volume": "886",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1000896",
"article-title": "Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing.",
"author": "Eckerle",
"doi-asserted-by": "publisher",
"journal-title": "PLoS Pathog.",
"key": "B9",
"volume": "6",
"year": "2010"
},
{
"DOI": "10.1007/978-1-4939-2438-7_1",
"article-title": "Coronaviruses: an overview of their replication and pathogenesis.",
"author": "Fehr",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Methods Mol. Biol.",
"key": "B10",
"volume": "1282",
"year": "2015"
},
{
"DOI": "10.1126/science.abb7498",
"article-title": "Structure of the RNA-dependent RNA polymerase from COVID-19 virus.",
"author": "Gao",
"doi-asserted-by": "publisher",
"first-page": "779",
"journal-title": "Science",
"key": "B11",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1128/JVI.02232-10",
"article-title": "Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response.",
"author": "Glowacka",
"doi-asserted-by": "publisher",
"first-page": "4122",
"journal-title": "J. Virol.",
"key": "B12",
"volume": "85",
"year": "2011"
},
{
"DOI": "10.1016/j.genrep.2020.100860",
"article-title": "In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors.",
"author": "Gurung",
"doi-asserted-by": "publisher",
"journal-title": "Gene Rep.",
"key": "B13",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1016/j.tmaid.2020.101646",
"article-title": "A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease.",
"author": "Hall",
"doi-asserted-by": "publisher",
"journal-title": "Travel Med. Infect. Dis.",
"key": "B14",
"volume": "35",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor.",
"author": "Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "271.e8",
"journal-title": "Cell",
"key": "B15",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1002/hpm.3006",
"article-title": "Known SARS-CoV-2 infections: the tip of an important iceberg.",
"author": "Hosein",
"doi-asserted-by": "publisher",
"first-page": "1270",
"journal-title": "Int. J. Health Plann. Manage.",
"key": "B16",
"volume": "35",
"year": "2020"
},
{
"DOI": "10.1128/JVI.78.11.5619-5632.2004",
"article-title": "Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase.",
"author": "Ivanov",
"doi-asserted-by": "publisher",
"first-page": "5619",
"journal-title": "J. Virol.",
"key": "B17",
"volume": "78",
"year": "2004"
},
{
"DOI": "10.1016/j.apsb.2020.04.009",
"article-title": "Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites.",
"author": "Kang",
"doi-asserted-by": "publisher",
"first-page": "1228",
"journal-title": "Acta Pharm. Sin. B",
"key": "B18",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.jiph.2020.06.016",
"article-title": "In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.",
"author": "Kumar",
"doi-asserted-by": "publisher",
"first-page": "1210",
"journal-title": "J. Infect. Public Health",
"key": "B19",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1021/ci500020m",
"article-title": "g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations.",
"author": "Kumari",
"doi-asserted-by": "publisher",
"first-page": "1951",
"journal-title": "J. Chem. Inf. Model",
"key": "B20",
"volume": "54",
"year": "2014"
},
{
"DOI": "10.1021/bi035923w",
"article-title": "Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines.",
"author": "Kwiek",
"doi-asserted-by": "publisher",
"first-page": "4538",
"journal-title": "Biochemistry",
"key": "B21",
"volume": "43",
"year": "2004"
},
{
"DOI": "10.21873/invivo.12134",
"article-title": "Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2.",
"author": "Lehrer",
"doi-asserted-by": "publisher",
"first-page": "3023",
"journal-title": "In Vivo",
"key": "B22",
"volume": "34",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1508686112",
"article-title": "Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex.",
"author": "Ma",
"doi-asserted-by": "publisher",
"first-page": "9436",
"journal-title": "Proc. Natl. Acad. Sci. U.S.A.",
"key": "B23",
"volume": "112",
"year": "2015"
},
{
"DOI": "10.1021/acs.jctc.5b00255",
"article-title": "ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB.",
"author": "Maier",
"doi-asserted-by": "publisher",
"first-page": "3696",
"journal-title": "J. Chem. Theory Comput.",
"key": "B24",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1016/j.virusres.2014.01.025",
"article-title": "Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities.",
"author": "Mielech",
"doi-asserted-by": "publisher",
"first-page": "184",
"journal-title": "Virus Res.",
"key": "B25",
"volume": "194",
"year": "2014"
},
{
"DOI": "10.1101/2020.06.26.174698",
"article-title": "A facile Q-RT-PCR assay for monitoring SARS-CoV-2 growth in cell culture.",
"author": "Mugisha",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv [Prepint]",
"key": "B26",
"year": "2020"
},
{
"journal-title": "First Antiviral Drug Approved to Fight Coronavirus. 2020.",
"key": "B27",
"year": "2020"
},
{
"DOI": "10.2210/pdb6w9c/pdb",
"article-title": "“The crystal structure of papain-like protease of SARS CoV-2,”",
"author": "Osipiuk",
"doi-asserted-by": "publisher",
"journal-title": "Center for Structural Genomics of Infectious Diseases (CSGID)",
"key": "B28",
"year": "2020"
},
{
"DOI": "10.1002/jcc.20084",
"article-title": "UCSF chimera–a visualization system for exploratory research and analysis.",
"author": "Pettersen",
"doi-asserted-by": "publisher",
"first-page": "1605",
"journal-title": "J. Comput. Chem.",
"key": "B29",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1128/JVI.79.17.11507-11512.2005",
"article-title": "Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells.",
"author": "Rowland",
"doi-asserted-by": "publisher",
"first-page": "11507",
"journal-title": "J. Virol.",
"key": "B30",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1006/jmbi.1993.1626",
"article-title": "Comparative protein modelling by satisfaction of spatial restraints.",
"author": "Sali",
"doi-asserted-by": "publisher",
"first-page": "779",
"journal-title": "J. Mol. Biol.",
"key": "B31",
"volume": "234",
"year": "1993"
},
{
"DOI": "10.1016/S1473-3099(06)70361-9",
"article-title": "New insights into the antiviral effects of chloroquine.",
"author": "Savarino",
"doi-asserted-by": "publisher",
"first-page": "67",
"journal-title": "Lancet Infect. Dis.",
"key": "B32",
"volume": "6",
"year": "2006"
},
{
"DOI": "10.1101/2020.05.15.098731",
"article-title": "Favipiravir strikes the SARS-CoV-2 at its achilles heel, the RNA polymerase.",
"author": "Shannon",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv [Preprint]",
"key": "B33",
"year": "2020"
},
{
"DOI": "10.1038/s41467-019-13940-6",
"article-title": "Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV.",
"author": "Sheahan",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "B34",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1128/JVI.01052-08",
"article-title": "The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles.",
"author": "Siu",
"doi-asserted-by": "publisher",
"first-page": "11318",
"journal-title": "J. Virol.",
"key": "B35",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1186/1756-0500-5-367",
"article-title": "ACPYPE - AnteChamber PYthon Parser interfacE.",
"author": "Sousa da Silva",
"doi-asserted-by": "publisher",
"journal-title": "BMC Res. Notes",
"key": "B36",
"volume": "5",
"year": "2012"
},
{
"DOI": "10.1016/j.antiviral.2013.06.002",
"article-title": "Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin.",
"author": "Tay",
"doi-asserted-by": "publisher",
"first-page": "301",
"journal-title": "Antiviral Res.",
"key": "B37",
"volume": "99",
"year": "2013"
},
{
"DOI": "10.1016/j.virusres.2005.05.007",
"article-title": "Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus.",
"author": "Timani",
"doi-asserted-by": "publisher",
"first-page": "23",
"journal-title": "Virus Res.",
"key": "B38",
"volume": "114",
"year": "2005"
},
{
"DOI": "10.1093/nar/gky1049",
"article-title": "UniProt: a worldwide hub of protein knowledge.",
"doi-asserted-by": "publisher",
"first-page": "D506",
"journal-title": "Nucleic Acids Res.",
"key": "B39",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1186/1743-422X-2-69",
"article-title": "Chloroquine is a potent inhibitor of SARS coronavirus infection and spread.",
"author": "Vincent",
"doi-asserted-by": "publisher",
"journal-title": "Virol. J.",
"key": "B40",
"volume": "2",
"year": "2005"
},
{
"DOI": "10.1042/BJ20120150",
"article-title": "Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus.",
"author": "Wagstaff",
"doi-asserted-by": "publisher",
"first-page": "851",
"journal-title": "Biochem. J.",
"key": "B41",
"volume": "443",
"year": "2012"
},
{
"DOI": "10.1016/j.cell.2020.11.032",
"article-title": "Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein.",
"author": "Walls",
"doi-asserted-by": "publisher",
"first-page": "281.e6",
"journal-title": "Cell",
"key": "B42",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.jmgm.2005.12.005",
"article-title": "Automatic atom type and bond type perception in molecular mechanical calculations.",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "247",
"journal-title": "J. Mol. Graph. Model",
"key": "B43",
"volume": "25",
"year": "2006"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro.",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "269",
"journal-title": "Cell Res.",
"key": "B44",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)31022-9",
"article-title": "Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial.",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1569",
"journal-title": "Lancet",
"key": "B45",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1038/nature17180",
"article-title": "Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys.",
"author": "Warren",
"doi-asserted-by": "crossref",
"first-page": "381",
"journal-title": "Nature",
"key": "B46",
"volume": "531",
"year": "2016"
},
{
"journal-title": "WHO Coronavirus Disease (COVID-19) Dashboard.",
"key": "B47",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.02.001",
"article-title": "Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China.",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "325",
"journal-title": "Cell Host Microbe",
"key": "B48",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.apsb.2020.02.008",
"article-title": "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods.",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "766",
"journal-title": "Acta Pharm. Sin. B",
"key": "B49",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkg916",
"article-title": "Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design.",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "7117",
"journal-title": "Nucleic Acids Res.",
"key": "B50",
"volume": "31",
"year": "2003"
},
{
"DOI": "10.1038/cr.2012.165",
"article-title": "Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model.",
"author": "Yan",
"doi-asserted-by": "crossref",
"first-page": "300",
"journal-title": "Cell Res.",
"key": "B51",
"volume": "23",
"year": "2013"
},
{
"DOI": "10.1016/j.antiviral.2020.104760",
"article-title": "The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer.",
"author": "Yang",
"doi-asserted-by": "publisher",
"journal-title": "Antiviral Res.",
"key": "B52",
"volume": "177",
"year": "2020"
},
{
"DOI": "10.1126/science.abc1560",
"article-title": "Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.",
"author": "Yin",
"doi-asserted-by": "publisher",
"first-page": "1499",
"journal-title": "Science",
"key": "B53",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1126/science.abb3405",
"article-title": "Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors.",
"author": "Zhang",
"doi-asserted-by": "publisher",
"first-page": "409",
"journal-title": "Science",
"key": "B54",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2012.04.011",
"article-title": "Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology.",
"author": "Zhao",
"doi-asserted-by": "publisher",
"first-page": "607",
"journal-title": "Cell Host Microbe",
"key": "B55",
"volume": "11",
"year": "2012"
},
{
"DOI": "10.1056/NEJMoa2001017",
"article-title": "A novel coronavirus from patients with pneumonia in China, 2019.",
"author": "Zhu",
"doi-asserted-by": "publisher",
"first-page": "727",
"journal-title": "N. Engl. J. Med.",
"key": "B56",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1099/0022-1317-81-4-853",
"article-title": "Virus-encoded proteinases and proteolytic processing in the Nidovirales.",
"author": "Ziebuhr",
"doi-asserted-by": "publisher",
"first-page": "853",
"journal-title": "J. Gen. Virol.",
"key": "B57",
"volume": "81",
"year": "2000"
}
],
"reference-count": 57,
"references-count": 57,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fmicb.2020.592908/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.3389/crossmark-policy",
"volume": "11"
}