In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction
et al., International Journal of Molecular Sciences, doi:10.3390/ijms242216392, Nov 2023
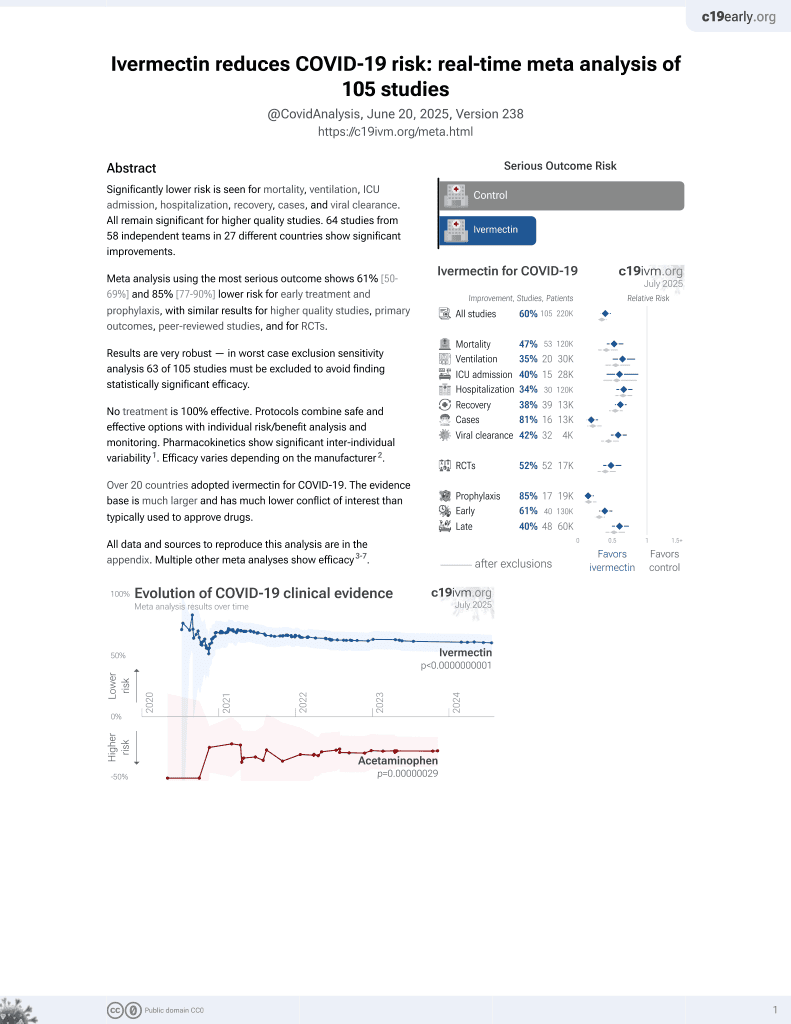
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
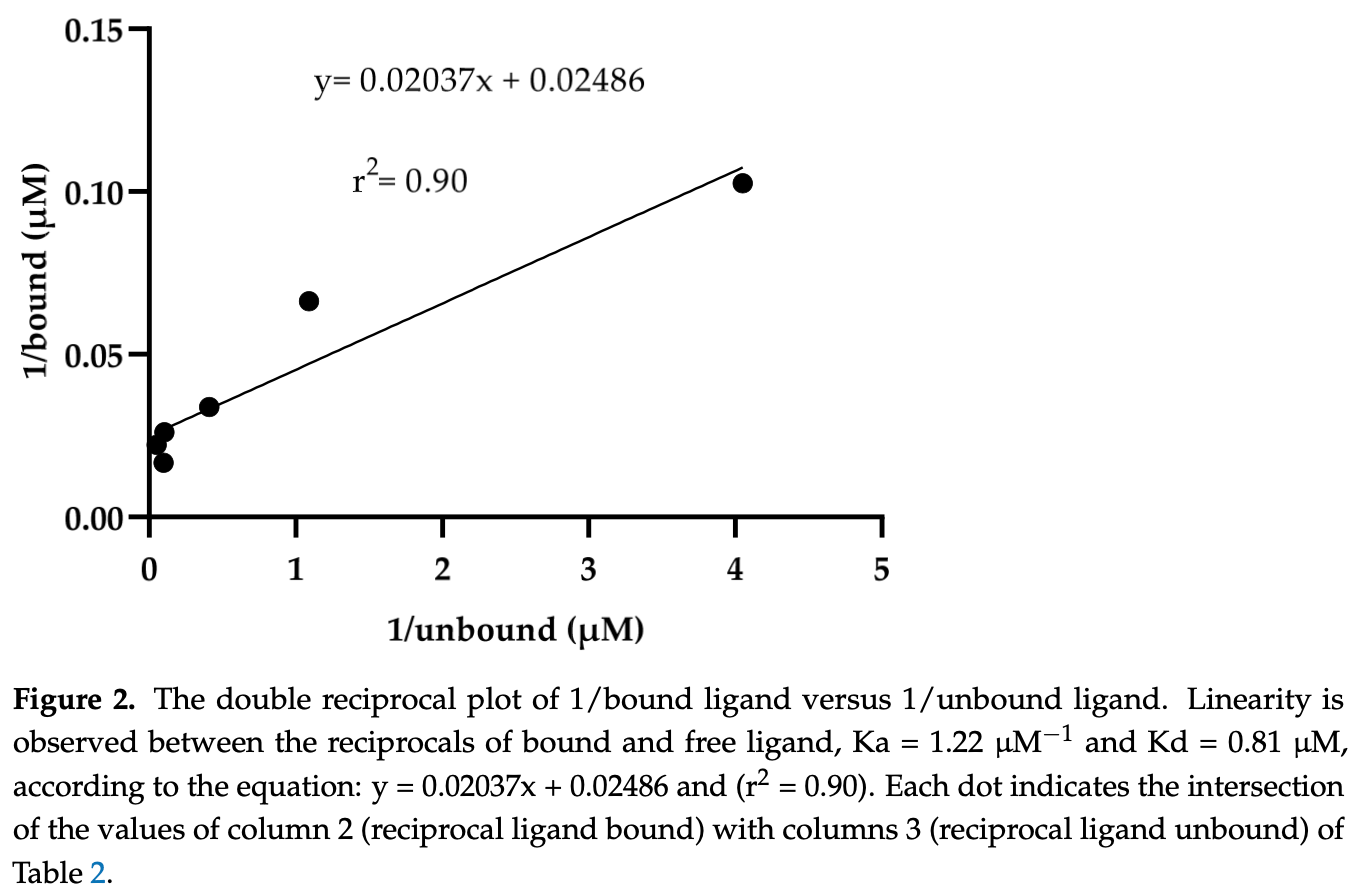
In vitro analysis showing a definitive interaction between ivermectin and the spike (S) protein of SARS-CoV-2, suggesting therapeutic potential for COVID-19. Using equilibrium dialysis and UV-Vis techniques, the study determined the affinity and dissociation constants between ivermectin and the S protein, finding an association constant (Ka) of 1.22 µM-1 and a dissociation constant (Kd) of 0.81 µM. Additionally, the Drug Affinity Responsive Target Stability (DARTS) method was employed, confirming the interaction in ratios of 1:50 pmol and 1:100 pmol (S: ivermectin).
Figure 2 shows the binding affinity curve between ivermectin and the spike protein S1/S2 fragment using equilibrium dialysis, showing the Ka and Kd values that indicate direct binding between ivermectin and spike. Figure 3 illustrates the Drug Affinity Responsive Target Stability (DARTS) gel showing protection of the spike protein from degradation when bound to ivermectin. This provides further evidence of direct binding between ivermectin and spike. Figure 4 compares the densitometry analysis of the DARTS gels, showing increased protection of spike protein at higher ivermectin doses. This supports the dose-dependent binding of ivermectin to spike.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
García-Aguilar et al., 16 Nov 2023, peer-reviewed, 7 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction
doi:10.3390/ijms242216392
The spike (S) protein of SARS-CoV-2 is a molecular target of great interest for developing drug therapies against COVID-19 because S is responsible for the interaction of the virus with the host cell receptor. Currently, there is no outpatient safety treatment for COVID-19 disease. Furthermore, we consider it of worthy importance to evaluate experimentally the possible interaction of drugs (approved by the Food and Drug Administration) and the S, considering some previously in silico and clinical use. Then, the objective of this study was to demonstrate the in vitro interaction of ivermectin with S. The equilibrium dialysis technique with UV-Vis was performed to obtain the affinity and dissociation constants. In addition, the Drug Affinity Responsive Target Stability (DARTS) technique was used to demonstrate the in vitro interaction of S with ivermectin. The results indicate the interaction between ivermectin and the S with an association and dissociation constant of Ka = 1.22 µM -1 and Kd = 0.81 µM, respectively. The interaction was demonstrated in ratios of 1:50 pmol and 1:100 pmol (S: ivermectin) by the DARTS technique. The results obtained with these two different techniques demonstrate an interaction between S and ivermectin previously explored in silico, suggesting its clinical uses to stop the viral spread among susceptible human hosts.
Supplementary Materials: The supporting information can be downloaded at: https://www.mdpi. com/article/10.3390/ijms242216392/s1.
Conflicts of Interest: The authors declare no conflict of interest.
References
Agostini, Andres, Sims, Graham, Sheahan et al., Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease, mBio, doi:10.1128/mBio.00221-18
Asselah, Durantel, Pasmant, Lau, Schinazi, COVID-19: Discovery, diagnostics and drug development, J. Hepatol, doi:10.1016/j.jhep.2020.09.031
Awad, Hassan, Dweek, Aboelata, Rawas-Qalaji et al., Repurposing Potential of the Antiparasitic Agent Ivermectin for the Treatment and/or Prophylaxis of COVID-19, Pharmaceuticals, doi:10.3390/ph15091068
Azam, Taban, Eid, Iqbal, Alam et al., An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin alpha, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1841028
Bell, Ligand Binding Module
Cevik, Grubaugh, Iwasaki, Openshaw, COVID-19 vaccines: Keeping pace with SARS-CoV-2 variants, Cell, doi:10.1016/j.cell.2021.09.010
Chaccour, Casellas, Blanco-Di Matteo, Pineda, Fernandez-Montero et al., The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: A pilot, double-blind, placebo-controlled, randomized clinical trial, EClinicalMedicine, doi:10.1016/j.eclinm.2020.100720
Choudhury, Das, Patra, Bhattacharya, Ghosh et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach, Future Virol, doi:10.2217/fvl-2020-0342
Clinicaltrials, Gov, Clinical Trial of Ivermectin Plus Doxycycline for the Treatment of Confirmed Covid-19 Infection
Clinicaltrials, Gov, Efficacy, Safety and Tolerability of Ivermectin in Subjects Infected with SARS-CoV-2 with or without Symptoms (SILVERBULLET)
Clinicaltrials, Gov, Ivermectin for Severe COVID-19 Management
Conterno, Turchi, Corrêa, Monteiro De Barros Almeida, Anthelmintic drugs for treating ascariasis, Cochrane Database Syst. Rev, doi:10.1002/14651858.CD010599.pub2
Correa-Basurto, Romero-Castro, Correa-Basurto, Hernandez-Rodriguez, Soriano-Ursua et al., Pharmacokinetics and tissue distribution of N-(2-hydroxyphenyl)-2-propylpentanamide in Wistar Rats and its binding properties to human serum albumin, J. Pharm. Biomed. Anal, doi:10.1016/j.jpba.2018.09.010
Deshpande, Tiwari, Nyayanit, Modak, In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2, Eur. J. Pharmacol, doi:10.1016/j.ejphar.2020.173430
Fda, Actualización Sobre El Coronavirus (COVID-19): La FDA Autoriza un Antiviral Oral Adicional
Food, Administration, Autorización de Uso de Emergencia de Paxlovid Para la Enfermedad por Coronavirus 2019
Gilead, European Commission Expands Indication for Veklury (Remdesivir) for the Treatment of Adults Not on Supplemental Oxygen and Considered High Risk for COVID-19 Disease Progression
Gonzalez, Gonzalez, Ueno, Ivermectin in Human Medicine, An Overview of the Current Status of Its Clinical Applications, Curr. Pharm. Biotechnol, doi:10.2174/138920112800399248
Gordon, Tchesnokov, Woolner, Perry, Feng et al., Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency, J. Biol. Chem, doi:10.1074/jbc.RA120.013679
Heidary, Gharebaghi, Ivermectin: A systematic review from antiviral effects to COVID-19 complementary regimen, J. Antibiot, doi:10.1038/s41429-020-0336-z
Huang, Yang, Xu, Xu, Liu, Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19, Acta Pharmacol. Sin, doi:10.1038/s41401-020-0485-4
Jans, Wagstaff, The broad spectrum host-directed agent ivermectin as an antiviral for SARS-CoV-2?, Biochem. Biophys. Res. Commun, doi:10.1016/j.bbrc.2020.10.042
Kim, COVID-19 Drug Development, J. Microbiol. Biotechnol, doi:10.4014/jmb.2110.10029
Kong, Chang, Qiao, Zhao, Chen et al., Paxlovid accelerates cartilage degeneration and senescence through activating endoplasmic reticulum stress and interfering redox homeostasis, J. Transl. Med, doi:10.1186/s12967-022-03770-4
Lehrer, Rheinstein, Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, Vivo, doi:10.21873/invivo.12134
Lomenick, Hao, Jonai, Chin, Aghajan et al., Target identification using drug affinity responsive target stability (DARTS), Proc. Natl. Acad. Sci, doi:10.1073/pnas.0910040106
Lomenick, Jung, Wohlschlegel, Huang, Target identification using drug affinity responsive target stability (DARTS), Curr. Protoc. Chem. Biol, doi:10.1002/9780470559277.ch110180
Lomenick, Olsen, Huang, Identification of direct protein targets of small molecules, ACS Chem. Biol, doi:10.1021/cb100294v
Luis, García, Unión a proteínas plasmáticas de la DL-3-hidroxi-3-etil-3-fenil-propionamida (HEPP). Un nuevo anticonvulsivante, J. Mex. Chem. Soc
Mali, Eerike, Raj, Bisoi, Priyadarshini et al., Efficacy and safety of Molnupiravir in COVID-19 patients: A systematic review, Ir. J. Med. Sci, doi:10.1007/s11845-022-03139-y
Malone, Campbell, Molnupiravir, Coding for catastrophe, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00657-8
Malone, Urakova, Snijder, Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00432-z
Mathachan, Sardana, Khurana, Current Use of Ivermectin in Dermatology, Tropical Medicine, and COVID-19: An Update on Pharmacology, Uses, Proven and Varied Proposed Mechanistic Action, Indian Dermatol. Online J
Merck, Merck and Ridgeback's Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study
Moctezuma, La glucoproteína spike, Rev. Mex. Mastol
Nafisi, Vishkaee, Study on the interaction of tamiflu and oseltamivir carboxylate with human serum albumin, J. Photochem. Photobiol. B, doi:10.1016/j.jphotobiol.2011.06.008
Niraj, Mahajan, Prakash, Sarma, Medhi et al., A promising drug for the challenging treatment of SARS-COV-2 in the pandemic era, Indian J. Pharmacol
Pfizer, Pfizer's Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 EPIC-HR Study
Polak, Van Gool, Cohen, Von Der Thusen, Van Paassen, A systematic review of pathological findings in COVID-19: A pathophysiological timeline and possible mechanisms of disease progression, Mod. Pathol, doi:10.1038/s41379-020-0603-3
Raman, Patel, Ranjan, COVID-19: Unmasking Emerging SARS-CoV-2 Variants, Vaccines and Therapeutic Strategies, Biomolecules, doi:10.3390/biom11070993
Ramirez-Salinas, Martinez-Archundia, Correa-Basurto, Garcia-Machorro, Repositioning of Ligands That Target the Spike Glycoprotein as Potential Drugs for SARS-CoV-2 in an In Silico Study, Molecules, doi:10.3390/molecules25235615
Reina, Iglesias, Nirmatrelvir plus ritonavir (Paxlovid) a potent SARS-CoV-2 3CLpro protease inhibitor combination, Rev. Esp. Quim, doi:10.37201/req/002.2022
Sanchezruiz, Nuzum, Kouzi, Oral ivermectin for the treatment of head lice infestation, Am. J. Health-Syst. Pharm, doi:10.2146/ajhp170464
Sanderson, Hisner, Donovan-Banfield, Hartman, Lochen et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6
Saravolatz, Depcinski, Sharma, Molnupiravir and Nirmatrelvir-Ritonavir: Oral Coronavirus Disease 2019 Antiviral Drugs, Clin. Infect. Dis, doi:10.1093/cid/ciac180
Sheppard, Laskou, Stapleton, Hadavi, Dasgupta, Tocilizumab (Actemra), Hum. Vaccin. Immunother, doi:10.1080/21645515.2017.1316909
Soufi, Iravani, Potential inhibitors of SARS-CoV-2: Recent advances, J. Drug Target, doi:10.1080/1061186X.2020.1853736
Tang, Bidon, Jaimes, Whittaker, Daniel, Coronavirus membrane fusion mechanism offers a potential target for antiviral development, Antivir. Res, doi:10.1016/j.antiviral.2020.104792
Tregoning, Flight, Higham, Wang, Pierce, Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape, Nat. Rev. Immunol, doi:10.1038/s41577-021-00592-1
Vuignier, Schappler, Veuthey, Carrupt, Martel, Drug-protein binding: A critical review of analytical tools, Anal. Bioanal. Chem, doi:10.1007/s00216-010-3737-1
Wang, Zhao, Gao, Gao, Wang et al., SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development, Front. Cell Infect. Microbiol, doi:10.3389/fcimb.2020.587269
Wen, Yan, Sun, Fang, Yang et al., A randomized, double-blind, multicenter clinical trial on the efficacy of ivermectin against intestinal nematode infections in China, Acta Trop, doi:10.1016/j.actatropica.2008.03.007
Zhou, Wang, Liu, Lu, Dong et al., Probing antiviral drugs against SARS-CoV-2 through virus-drug association prediction based on the KATZ method, Genomics, doi:10.1016/j.ygeno.2020.07.044