A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes
et al., Nature, doi:10.1038/s41586-023-06649-6, Sep 2023
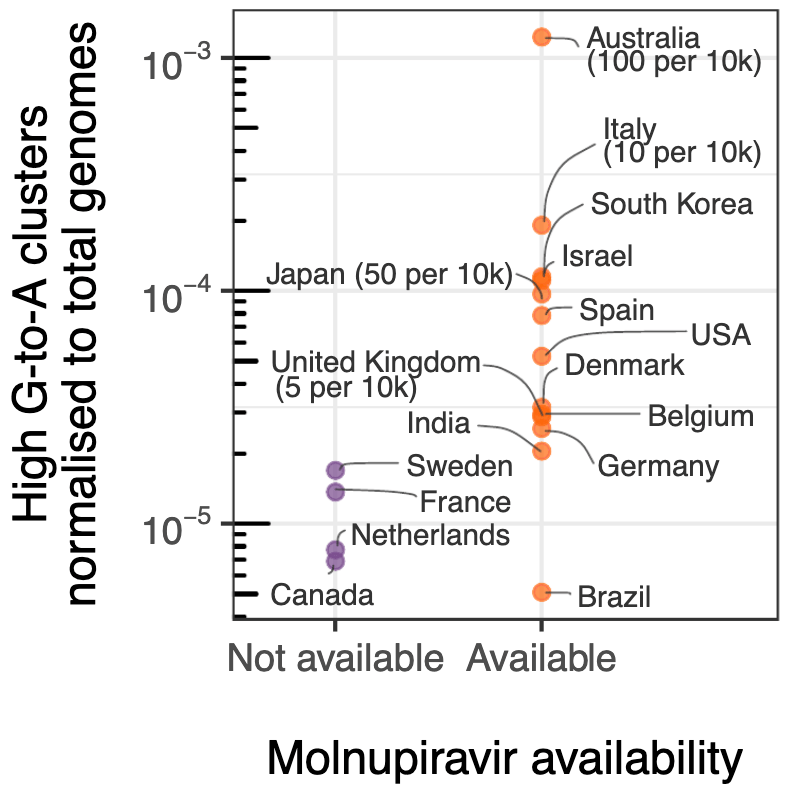
Identification of SARS-CoV-2 variants created by molnupiravir treatment, including cases of onwards transmission. Authors find a class of long phylogenetic branches almost exclusively matching the time period, location, and age groups of widespread molnupiravir treatment. There were extreme cases with >100 molnupiravir-associated mutations, and confirmed cases where molnupiravir-derived sequences were transmitted to others. For more discussion see1.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity2-16. Multiple analyses have identified variants potentially created by molnupiravir17-21. Studies show significantly increased risk of acute kidney injury22, cardiovascular toxocity23, and neurological symptoms22. Treatment may increase viral rebound24,25.
2.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
3.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
4.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
5.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
6.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
7.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
8.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
9.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
10.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
11.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
12.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
13.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
14.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
15.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
16.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
17.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
18.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
19.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
20.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
22.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
23.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Sanderson et al., 25 Sep 2023, peer-reviewed, 7 authors.
Contact: theo.sanderson@crick.ac.uk (corresponding author), cr628@cam.ac.uk.
Identification of a molnupiravir-associated mutational signature in SARS-CoV-2 sequencing databases
doi:10.1101/2023.01.26.23284998
Molnupiravir, an antiviral medication that has been widely used against SARS-CoV-2, acts by inducing mutations in the virus genome during replication. Most random mutations are likely to be deleterious to the virus, and many will be lethal. Molnupiravirinduced elevated mutation rates have been shown to decrease viral load in animal models. However, it is possible that some patients treated with molnupiravir might not fully clear SARS-CoV-2 infections, with the potential for onward transmission of molnupiravir-mutated viruses. We set out to systematically investigate global sequencing databases for a signature of molnupiravir mutagenesis. We find that a specific class of long phylogenetic branches appear almost exclusively in sequences from 2022, after the introduction of molnupiravir treatment, and in countries and agegroups with widespread usage of the drug. We calculate a mutational spectrum from the AGILE placebo-controlled clinical trial of molnupiravir and show that its signature, with elevated G-to-A and C-to-T rates, largely corresponds to the mutational spectrum seen in these long branches. Our data suggest a signature of molnupiravir mutagenesis can be seen in global sequencing databases, in some cases with onwards transmission.
AUTHOR CONTRIBUTIONS RH identified initial branches, and their likely connection to molnupiravir. TS performed analyses of mutation-annotated tree and global metadata. CR performed all mutational spectra analyses. ID-B created bioinformatic pipelines for the AGILE trial data. All authors participated in mansuscript writing.
Supplementary Information
T
Figure S1. Possible outcomes from MTP incorporation This figure depicts some of the mutational pathways related to MTP incorporation into MTP. The first column shows what may be a common event, but is not detectable by sequencing. MTP can be incorporated into RNA (pairing with G) and then pair with G again in the next round of synthesis, which will result in no mutation in the final sequence. However if the MTP takes on an alternative tautomeric form after incorporation it can bind to A, creating a G-to-A mutation. The third column shows that if the positive-sense base is C, then this will bind to a G in the formation of the negative-sense genome. In subsequent replication this negative sense genome can undergo the same G-to-A mutation seen in the second column, which ultimately results in a positive sense C-to-T mutation. Although the biases of tautomeric forms for the free and incorporated MTP nucleotides appear to favour these directionalities of mutations, the reverse is also possible, resulting in A-to-G and T-to-C mutations.
Figure S2. High G-to-A branches involve the same number of mutations occurring in a..
References
Aksamentov, Roemer, Hodcroft, Neher, Nextclade: clade assignment, mutation calling and quality control for viral genomes, Journal of Open Source Software
Bernal, Gomes Da Silva, Musungaie, Kovalchuk, Gonzalez et al., Molnupiravir for oral treatment of covid-19 in nonhospitalized patients, N. Engl. J. Med
Bloom, Beichman, Neher, Harris, Evolution of the SARS-CoV-2 mutational spectrum
Butler, Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial, Lancet
Carabelli, Peacock, Thorne, Harvey, Hughes et al., Sars-CoV-2 variant biology: immune escape, transmission and fitness, Nature Reviews Microbiology
Cochrane, Karsch-Mizrachi, Nakamura, and on behalf of the International Nucleotide Sequence Database Collaboration
Donovan-Banfield, Penrice-Randal, Goldswain, Rzeszutek, Pilgrim et al., Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial, Nat Commun
Eigen, Selforganization of matter and the evolution of biological macromolecules, Naturwissenschaften
Elbe, Buckland-Merrett, Data, disease and diplomacy: GISAID's innovative contribution to global health, Glob Chall
Extance, Covid-19: What is the evidence for the antiviral molnupiravir?, BMJ
Fountain-Jones, Vanhaeften, Williamson, Maskell, Chua et al., Antiviral treatments lead to the rapid accrual of hundreds of SARS-CoV-2 mutations in immunocompromised patients
Gold, Kelleher, Magid, Jackson, Pennini et al., Dispensing of Oral Antiviral Drugs for Treatment of COVID-19 by Zip Code-Level Social Vulnerability -United States, MMWR Morb Mortal Wkly Rep
Gordon, Tchesnokov, Schinazi, Götte, Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template
Hadfield, Megill, Bell, Huddleston, Potter et al., Nextstrain: real-time tracking of pathogen evolution, Bioinformatics
Harari, Tahor, Rutsinsky, Meijer, Miller et al., Drivers of adaptive evolution during chronic SARS-CoV-2 infections, Nat Med
Hill, Du Plessis, Peacock, Aggarwal, Colquhoun et al., The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK, Virus Evol
Hisner, Re, Potential BA.2.3 sublineage with many mutations (singleton, Indonesia
Khoo, Fitzgerald, Saunders, Middleton, Ahmad et al., Molnupiravir versus placebo in unvaccinated and vaccinated patients with early SARS-CoV-2 infection in the UK (AGILE CST-2): a randomised, placebo-controlled, double-blind, phase 2 trial, Lancet Infect Dis
Malone, Campbell, Molnupiravir: coding for catastrophe, Nat Struct Mol Biol
Masone, Alvarez, Polo, The SARS-CoV-2 mutation landscape is shaped before replication starts
Mcbroome, Thornlow, Hinrichs, Kramer, De Maio et al., A Daily-Updated database and tools for comprehensive SARS-CoV-2 Mutation-Annotated trees, Mol. Biol. Evol
Minh, Schmidt, Chernomor, Schrempf, Woodhams et al., Iq-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era, Molecular Biology and Evolution
Nice Guidance, NICE recommends 3 treatments for COVID-19
Rambaut, Figtree, None
Rambaut, Loman, Pybus, Barclay, Barrett et al., Preliminary genomic characterisation of an emergent sars-cov-2 lineage in the uk defined by a novel set of spike mutations
Reuters ; Rosenke, Hansen, Schwarz, Feldmann, Haddock et al., Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the syrian hamster model, Nat. Commun
Ruis, Peacock, Polo, Masone, Alvarez et al., Mutational spectra distinguish SARS-CoV-2 replication niches
Sagulenko, Puller, Neher, Treetime: Maximum-likelihood phylodynamic analysis, Virus evolution
Sanderson, Biorxiv, None
Sanderson, Taxonium, Spencer, a web-based tool for exploring large phylogenetic trees
Summers, Litwin, Examining the theory of error catastrophe, J. Virol
Telenti, Hodcroft, Robertson, The evolution and biology of SARS-CoV-2 variants, Cold Spring Harbor Perspectives in Medicine
Tonkin-Hill, Martincorena, Amato, Lawson, Gerstung et al., Patterns of within-host genetic diversity in SARS-CoV-2
Turakhia, Thornlow, Hinrichs, De Maio, Gozashti et al., Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic, Nat Genet
Viana, Moyo, Amoako, Tegally, Scheepers et al., Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa, Nature
Wirth, Duchene, Gisaidr, None, doi:10.5281/zenodo.6474693
DOI record:
{
"DOI": "10.1038/s41586-023-06649-6",
"ISSN": [
"0028-0836",
"1476-4687"
],
"URL": "http://dx.doi.org/10.1038/s41586-023-06649-6",
"alternative-id": [
"6649"
],
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "27 January 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "15 September 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "25 September 2023"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-4177-2851",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sanderson",
"given": "Theo",
"sequence": "first"
},
{
"affiliation": [],
"family": "Hisner",
"given": "Ryan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5124-2427",
"affiliation": [],
"authenticated-orcid": false,
"family": "Donovan-Banfield",
"given": "I’ah",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hartman",
"given": "Hassan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Løchen",
"given": "Alessandra",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7077-2928",
"affiliation": [],
"authenticated-orcid": false,
"family": "Peacock",
"given": "Thomas P.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0977-5534",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ruis",
"given": "Christopher",
"sequence": "additional"
}
],
"container-title": "Nature",
"container-title-short": "Nature",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2023,
9,
25
]
],
"date-time": "2023-09-25T16:05:02Z",
"timestamp": 1695657902000
},
"deposited": {
"date-parts": [
[
2023,
9,
25
]
],
"date-time": "2023-09-25T16:12:21Z",
"timestamp": 1695658341000
},
"indexed": {
"date-parts": [
[
2023,
9,
26
]
],
"date-time": "2023-09-26T05:53:25Z",
"timestamp": 1695707605157
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
9,
25
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.springernature.com/gp/researchers/text-and-data-mining",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
25
]
],
"date-time": "2023-09-25T00:00:00Z",
"timestamp": 1695600000000
}
},
{
"URL": "https://www.springernature.com/gp/researchers/text-and-data-mining",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
25
]
],
"date-time": "2023-09-25T00:00:00Z",
"timestamp": 1695600000000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41586-023-06649-6.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41586-023-06649-6",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41586-023-06649-6.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2023,
9,
25
]
]
},
"published-online": {
"date-parts": [
[
2023,
9,
25
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41586-023-06649-6"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Multidisciplinary"
],
"subtitle": [],
"title": "A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy"
}