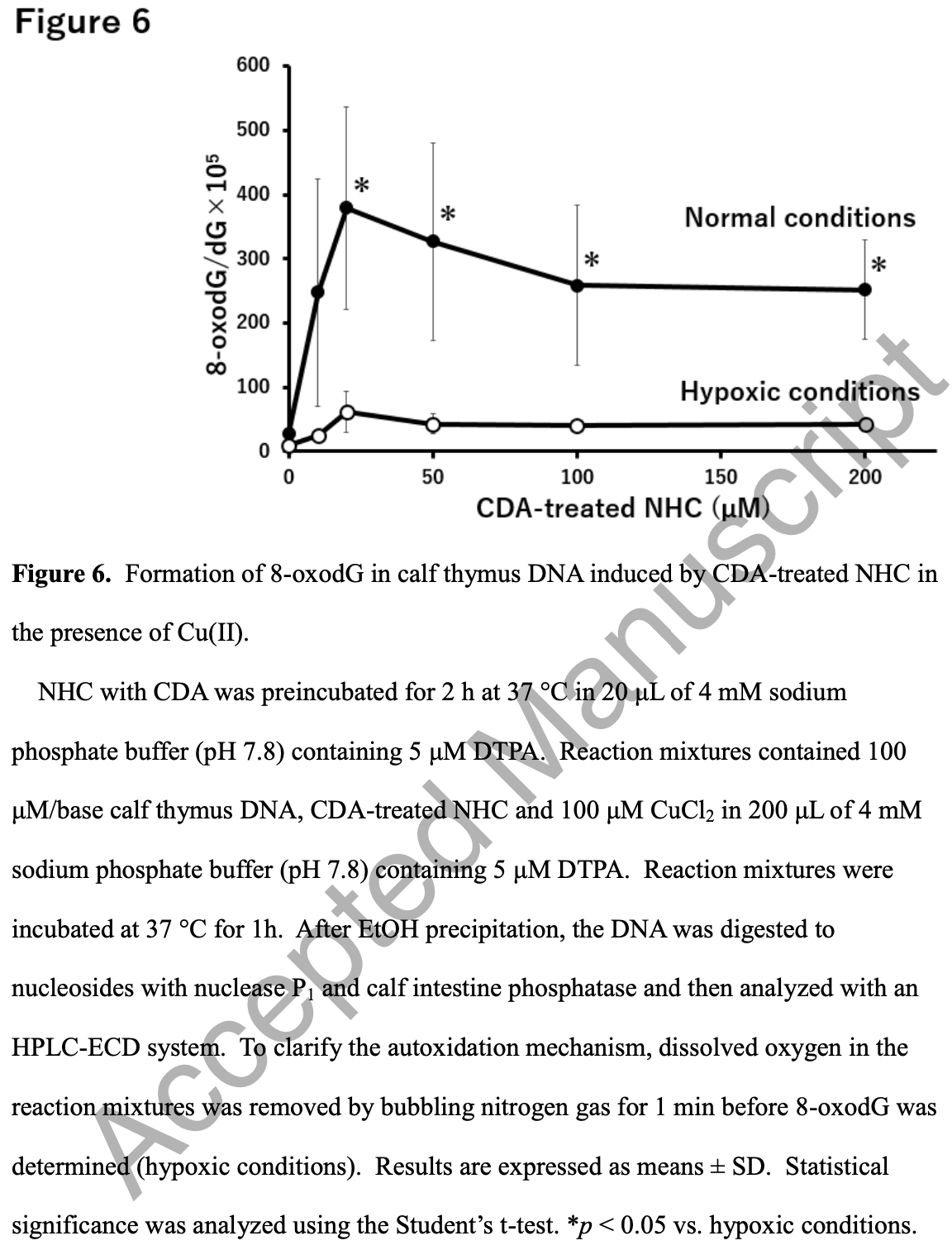
Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir
et al., Free Radical Research, doi:10.1080/10715762.2025.2469738, Feb 2025
In vitro study showing that molnupiravir may have cytotoxic and mutagenic effects in host cells via hydroxylamine production from N4-hydroxycytidine (NHC) by cytidine deaminase (CDA). Molnupiravir metabolite NHC may induce cytotoxicity and mutagenicity through CDA-mediated reactive oxygen species generation.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Mori et al., 20 Feb 2025, Japan, peer-reviewed, 10 authors.
Contact: s-oikawa@med.mie-u.ac.jp.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: Free Radical Research
ISSN: (Print) (Online) Journal homepage: www.tandfonline.com/journals/ifra20
Reactive oxygen species-mediated cytotoxic and
DNA-damaging mechanism of N4-hydroxycytidine,
a metabolite of the COVID-19 therapeutic drug
molnupiravir
Yurie Mori, Rinya Yogo, Hatasu Kobayashi, Hirotaka Katsuzaki, Yuichiro
Hirao, Shinya Kato, Hirokazu Kotani, Shosuke Kawanishi, Mariko Murata &
Shinji Oikawa
To cite this article: Yurie Mori, Rinya Yogo, Hatasu Kobayashi, Hirotaka Katsuzaki, Yuichiro
Hirao, Shinya Kato, Hirokazu Kotani, Shosuke Kawanishi, Mariko Murata & Shinji Oikawa (20
4
Feb 2025): Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical
Research, DOI: 10.1080/10715762.2025.2469738
To link to this article: https://doi.org/10.1080/10715762.2025.2469738
© 2025 The Author(s). Published by Informa
UK Limited, trading as Taylor & Francis
Group
Accepted author version posted online: 20
Feb 2025.
Submit your article to this journal
View related articles
View Crossmark data
Full Terms & Conditions of access and use can be found at
https://www.tandfonline.com/action/journalInformation?journalCode=ifra20
Revised manuscript
Reactive oxygen species-mediated cytotoxic and DNA-damaging
mechanism of N4-hydroxycytidine, a metabolite of the COVID-19
ip
t
therapeutic drug molnupiravir
Yurie Mori1,#, Rinya Yogo1,2,#, Hatasu Kobayashi1, Hirotaka Katsuzaki3, Yuichiro
cr
Hirao1, Shinya Kato4, Hirokazu Kotani 2, Shosuke Kawanishi5, Mariko Murata1, Shinji
an
Yurie Mori and Rinya Yogo should be considered joint first authors.
M
#
us
Oikawa1*
1. Department of Environmental and Molecular Medicine, Mie University Graduate
ed
School of Medicine, Edobashi 2-174, Tsu, Mie 514-8507, Japan
pt
2. Department of Forensic Medicine and Sciences, Mie University Graduate School of
ce
Medicine, Edobashi 2-174, Tsu, Mie 514-8507, Japan
3. Department of Life Sciences, Graduate School of Bioresources, Mie University,
Ac
1577 Kurimamachiya, Tsu, Mie 514-8507, Japan
4. Radioisotope Experimental Facility, Advanced Science Research Promotion Center,
Mie University, Edobashi 2-174, Tsu, Mie 514-8507, Japan
5. Faculty of Pharmaceutical Sciences, Suzuka University of Medical Science, 3500-3,
Minamitamagaki, Suzuka, Mie, 513-8670, Japan
1
* Corresponding author at: Department of Environmental and Molecular Medicine, Mie
University Graduate School of Medicine, Edobashi 2-174, Tsu, Mie 514-8507, Japan.
E-mail address: s-oikawa@med.mie-u.ac.jp (S. Oikawa)
Running title: ROS generation mechanism of hydroxycytidine
Keywords: molnupiravir, N4-hydroxycytidine, cytidine deaminase, reactive oxygen
Ac
ce
pt
ed
M
an
us
cr
ip
t
species, cytotoxicity, DNA damage
2
Abstract
Molnupiravir is a prodrug of the antiviral ribonucleoside analogue N4-hydroxycytidine
(NHC), for use in treatment of coronavirus disease 2019 (COVID-19). However, it is
generally considered that NHC-triphosphate is incorporated into the host genome to
ip
t
induce mutations. In our previous preliminary report, we proposed oxidative DNA
cr
damage by NHC via cytidine deaminase (CDA)-mediated ROS formation. In the
us
present study, we investigated cell viability using the HL-60 human leukemia cell line
an
and its H2O2-resistant clone, HP100 cells. The survival rate was significantly reduced
in HL-60 cells treated with NHC, but not in HP100 cells. LC-MS..
DOI record:
{
"DOI": "10.1080/10715762.2025.2469738",
"ISSN": [
"1071-5762",
"1029-2470"
],
"URL": "http://dx.doi.org/10.1080/10715762.2025.2469738",
"alternative-id": [
"10.1080/10715762.2025.2469738"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=ifra20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=ifra20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-08-21"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2024-12-05"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-01-13"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-02-20"
}
],
"author": [
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Mori",
"given": "Yurie",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
},
{
"name": "Department of Forensic Medicine and Sciences, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Yogo",
"given": "Rinya",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Kobayashi",
"given": "Hatasu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Life Sciences, Graduate School of Bioresources, Mie University",
"place": [
"Tsu, Japan"
]
}
],
"family": "Katsuzaki",
"given": "Hirotaka",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Hirao",
"given": "Yuichiro",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Radioisotope Experimental Facility, Advanced Science Research Promotion Center, Mie University",
"place": [
"Tsu, Japan"
]
}
],
"family": "Kato",
"given": "Shinya",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Forensic Medicine and Sciences, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Kotani",
"given": "Hirokazu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Faculty of Pharmaceutical Sciences, Suzuka University of Medical Science",
"place": [
"Minamitamagaki, Suzuka, Japan"
]
}
],
"family": "Kawanishi",
"given": "Shosuke",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Murata",
"given": "Mariko",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine",
"place": [
"Tsu, Japan"
]
}
],
"family": "Oikawa",
"given": "Shinji",
"sequence": "additional"
}
],
"container-title": "Free Radical Research",
"container-title-short": "Free Radical Research",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2025,
2,
20
]
],
"date-time": "2025-02-20T08:19:35Z",
"timestamp": 1740039575000
},
"deposited": {
"date-parts": [
[
2025,
2,
20
]
],
"date-time": "2025-02-20T08:19:54Z",
"timestamp": 1740039594000
},
"indexed": {
"date-parts": [
[
2025,
2,
21
]
],
"date-time": "2025-02-21T05:17:48Z",
"timestamp": 1740115068382,
"version": "3.37.3"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
2,
20
]
]
},
"language": "en",
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/10715762.2025.2469738",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "1-34",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2025,
2,
20
]
]
},
"published-online": {
"date-parts": [
[
2025,
2,
20
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1016/j.ijantimicag.2024.107096",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_2_1",
"unstructured": "Zheng B. et al. Small-molecule antiviral treatments for COVID-19: A systematic review and network meta-analysis. Int J Antimicrob Agents 2024. 63(3): p. 107096."
},
{
"DOI": "10.1126/scitranslmed.aax5866",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_3_1",
"unstructured": "Toots M. et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med 2019. 11(515)."
},
{
"DOI": "10.1126/scitranslmed.abb5883",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_4_1",
"unstructured": "Sheahan T.P. et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 2020. 12(541)."
},
{
"DOI": "10.1073/pnas.1718806115",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_5_1",
"unstructured": "Ferron F. et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A 2018. 115(2): p. E162-E171."
},
{
"DOI": "10.3389/fimmu.2023.1125246",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_6_1",
"unstructured": "Yuan Y. et al. The development of COVID-19 treatment. Front Immunol 2023. 14: p. 1125246."
},
{
"DOI": "10.1136/bmj.o926",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_7_1",
"unstructured": "Extance A. Covid-19: What is the evidence for the antiviral molnupiravir? Bmj 2022. 377: p. o926."
},
{
"DOI": "10.1093/infdis/jiab247",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_8_1",
"unstructured": "Zhou S. et al. β-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells. J Infect Dis 2021. 224(3): p. 415-419."
},
{
"DOI": "10.1002/em.22471",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_9_1",
"unstructured": "Waters M.D. et al. Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: The special case of molnupiravir. Environ Mol Mutagen 2022. 63(1): p. 37-63."
},
{
"DOI": "10.1128/AAC.01719-19",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_10_1",
"unstructured": "Sticher Z.M. et al. Analysis of the Potential for N(4)-Hydroxycytidine To Inhibit Mitochondrial Replication and Function. Antimicrob Agents Chemother 2020. 64(2)."
},
{
"DOI": "10.1016/j.toxlet.2022.04.002",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_11_1",
"unstructured": "Brandsma I. et al. Genotoxicity assessment of potentially mutagenic nucleoside analogues using ToxTracker®. Toxicol Lett 2022. 362: p. 50-58."
},
{
"DOI": "10.1093/infdis/jiac477",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_12_1",
"unstructured": "Kobayashi H. et al. Oxidative DNA Damage by N4-hydroxycytidine a Metabolite of the SARS-CoV-2 Antiviral Molnupiravir. J Infect Dis 2023. 227(9): p. 1068-1072."
},
{
"DOI": "10.1093/carcin/7.11.1849",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_13_1",
"unstructured": "Kasai H. et al. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 1986. 7(11): p. 1849-51."
},
{
"DOI": "10.1289/ehp.99107469",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_14_1",
"unstructured": "Calderon-Garciduenas L. et al. 8-hydroxy-2'-deoxyguanosine a major mutagenic oxidative DNA lesion and DNA strand breaks in nasal respiratory epithelium of children exposed to urban pollution. Environ Health Perspect 1999. 107(6): p. 469-74."
},
{
"DOI": "10.1016/0168-9525(93)90089-Z",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_15_1",
"unstructured": "Grollman A.P. and M. Moriya Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet 1993. 9(7): p. 246-9."
},
{
"DOI": "10.1016/S0021-9258(18)48474-8",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_16_1",
"unstructured": "Cheng K.C. et al. 8-Hydroxyguanine an abundant form of oxidative DNA damage causes G––T and A––C substitutions. J Biol Chem 1992. 267(1): p. 166-72."
},
{
"DOI": "10.1038/366704a0",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_17_1",
"unstructured": "Serrano M. G.J. Hannon and D. Beach A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993. 366(6456): p. 704-7."
},
{
"DOI": "10.1038/302033a0",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_18_1",
"unstructured": "Capon D.J. et al. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature 1983. 302(5903): p. 33-7."
},
{
"DOI": "10.1038/sj.onc.1206440",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_19_1",
"unstructured": "Oikawa S. K. Murakami and S. Kawanishi Oxidative damage to cellular and isolated DNA by homocysteine: implications for carcinogenesis. Oncogene 2003. 22(23): p. 3530-8."
},
{
"DOI": "10.1111/j.1349-7006.2002.tb01314.x",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_20_1",
"unstructured": "Ohnishi S. M. Murata and S. Kawanishi Oxidative DNA damage induced by a metabolite of 2-naphthylamine a smoking-related bladder carcinogen. Jpn J Cancer Res 2002. 93(7): p. 736-43."
},
{
"DOI": "10.1080/10715762.2018.1562179",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_21_1",
"unstructured": "Mori Y. et al. Mechanisms of DNA damage induced by morin an inhibitor of amyloid beta-peptide aggregation. Free Radic Res 2019. 53(1): p. 115-123."
},
{
"DOI": "10.1016/0145-2126(92)90129-U",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_22_1",
"unstructured": "Kasugai I. and M. Yamada High production of catalase in hydrogen peroxide-resistant human leukemia HL-60 cell lines. Leuk Res 1992. 16(2): p. 173-9."
},
{
"DOI": "10.1016/S0076-6879(80)65059-9",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_23_1",
"unstructured": "Maxam A.M. and W. Gilbert Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 1980. 65(1): p. 499-560."
},
{
"DOI": "10.1016/0006-291X(78)91562-0",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_24_1",
"unstructured": "Pryor W.A. and R.H. Tang Ethylene formation from methional. Biochem Biophys Res Commun 1978. 81(2): p. 498-503."
},
{
"DOI": "10.1007/s002800000223",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_25_1",
"unstructured": "Gourdeau H. et al. Comparative study of a novel nucleoside analogue (Troxatyl troxacitabine BCH-4556) and AraC against leukemic human tumor xenografts expressing high or low cytidine deaminase activity. Cancer Chemother Pharmacol 2001. 47(3): p. 236-40."
},
{
"DOI": "10.1128/AAC.02428-20",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_26_1",
"unstructured": "Painter W.P. et al. Human Safety Tolerability and Pharmacokinetics of Molnupiravir a Novel Broad-Spectrum Oral Antiviral Agent with Activity Against SARS-CoV-2. Antimicrob Agents Chemother 2021. 65(5)."
},
{
"DOI": "10.1517/17425255.2015.985648",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_27_1",
"unstructured": "Serdjebi C. G. Milano and J. Ciccolini Role of cytidine deaminase in toxicity and efficacy of nucleosidic analogs. Expert Opin Drug Metab Toxicol 2015. 11(5): p. 665-72."
},
{
"DOI": "10.1128/mSphere.00374-19",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_28_1",
"unstructured": "Moro-Bulnes A. et al. Contribution of Cytidine Deaminase to Thymidylate Biosynthesis in Trypanosoma brucei: Intracellular Localization and Properties of the Enzyme. mSphere 2019. 4(4)."
},
{
"DOI": "10.1038/srep18191",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_29_1",
"unstructured": "Marx A. M. Galilee and A. Alian Zinc enhancement of cytidine deaminase activity highlights a potential allosteric role of loop-3 in regulating APOBEC3 enzymes. Sci Rep 2015. 5: p. 18191."
},
{
"DOI": "10.1021/bi00014a003",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_30_1",
"unstructured": "Xiang S. et al. Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase. Biochemistry 1995. 34(14): p. 4516-23."
},
{
"DOI": "10.3109/10408448509023765",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_31_1",
"unstructured": "Gross P. Biologic activity of hydroxylamine: a review. Crit Rev Toxicol 1985. 14(1): p. 87-99."
},
{
"DOI": "10.1007/978-1-4615-5381-6_60",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_32_1",
"unstructured": "Vincenzetti S. et al. Studies on cysteine residues involved in the active site of human cytidine deaminase. Adv Exp Med Biol 1998. 431: p. 305-8."
},
{
"DOI": "10.3390/ijms22094758",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_33_1",
"unstructured": "Maj P. et al. Molecular Mechanism of Thymidylate Synthase Inhibition by N(4)-Hydroxy-dCMP in View of Spectrophotometric and Crystallographic Studies. Int J Mol Sci 2021. 22(9)."
},
{
"DOI": "10.1085/jgp.7.3.373",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_34_1",
"unstructured": "Northrop J.H. The Kinetics of the Decomposition of Peroxide by Catalase. J Gen Physiol 1925. 7(3): p. 373-87."
},
{
"DOI": "10.1016/S0003-2697(02)00031-3",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_35_1",
"unstructured": "Rapisarda V.A. et al. Quenching of bathocuproine disulfonate fluorescence by Cu(I) as a basis for copper quantification. Anal Biochem 2002. 307(1): p. 105-9."
},
{
"DOI": "10.1186/s41021-023-00258-5",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_36_1",
"unstructured": "Toyokuni S. et al. Environmental impact on carcinogenesis under BRCA1 haploinsufficiency. Genes Environ 2023. 45(1): p. 2."
},
{
"DOI": "10.1016/j.cbi.2022.110173",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_37_1",
"unstructured": "Jomova K. et al. Essential metals in health and disease. Chem Biol Interact 2022. 367: p. 110173."
},
{
"DOI": "10.1186/s12199-018-0740-1",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_38_1",
"unstructured": "Murata M. Inflammation and cancer. Environ Health Prev Med 2018. 23(1): p. 50."
},
{
"DOI": "10.1016/S0891-5849(02)00969-3",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_39_1",
"unstructured": "Sakano K. et al. Metabolism of carcinogenic urethane to nitric oxide is involved in oxidative DNA damage. Free Radic Biol Med 2002. 33(5): p. 703-14."
}
],
"reference-count": 38,
"references-count": 38,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/10715762.2025.2469738"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of\n <i>N</i>\n <sup>4</sup>\n -hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir",
"type": "journal-article",
"update-policy": "https://doi.org/10.1080/tandf_crossmark_01"
}