Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients
et al., Nature Communications, doi:10.1038/s41467-024-45641-0, ISRCTN30448031, Feb 2024
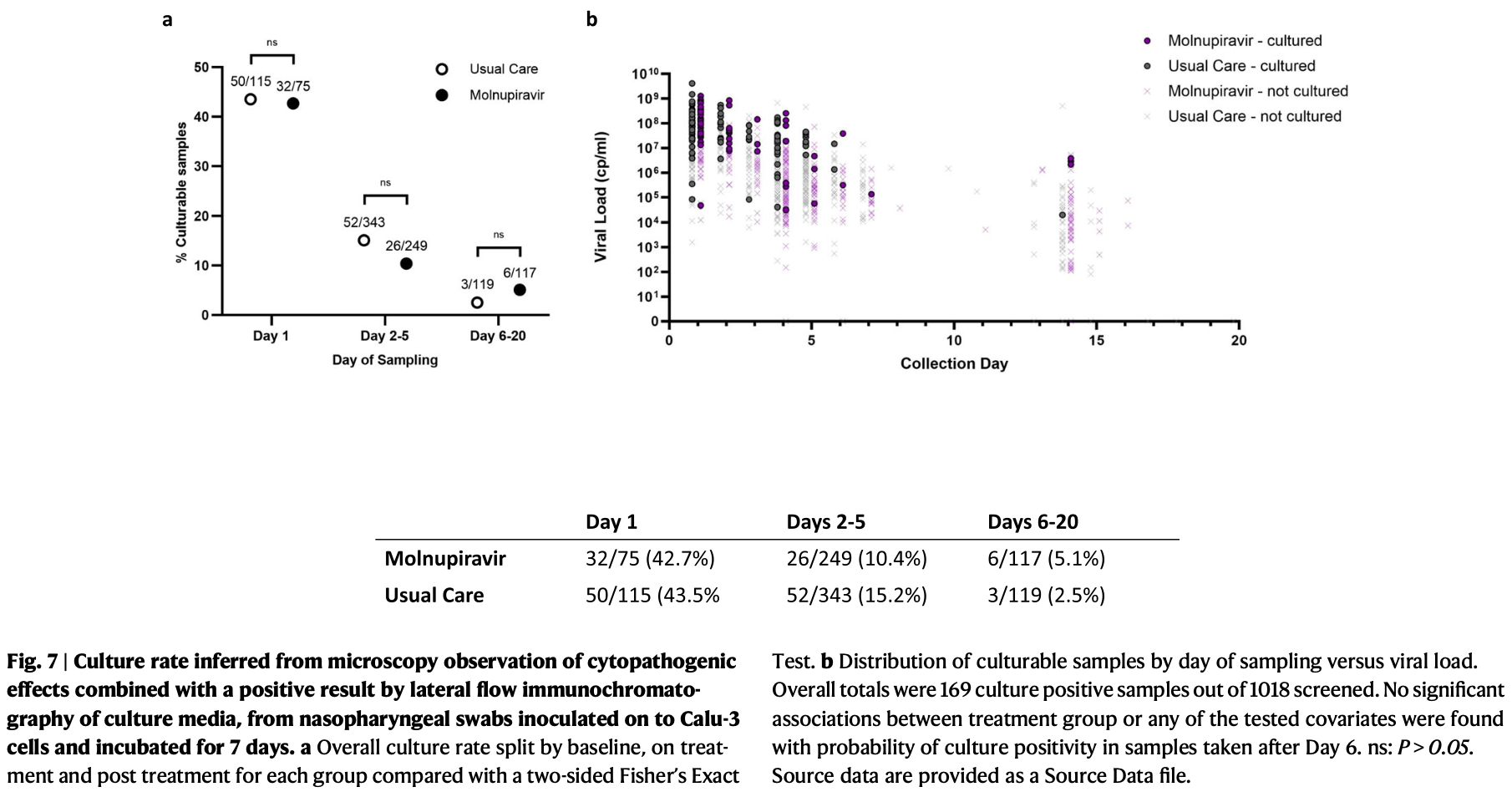
PANORAMIC virology-sub-study showing increased viral persistence with molnupiravir treatment. Molnupiravir 800mg twice daily for 5 days led to faster initial viral decline but 86% still had detectable virus by day 5. By day 14, molnupiravir was associated with significantly higher proportions with detectable virus and lower anti-spike antibodies compared to usual care. Serial genome sequencing revealed substantially increased mutagenesis with molnupiravir. Viable virus was cultured from samples up to 9 days post-treatment.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
Standard of Care (SOC) for COVID-19 in the study country,
the United Kingdom, is very poor with very low average efficacy for approved treatments25.
The United Kingdom focused on expensive high-profit treatments, approving only one low-cost early treatment, which required a prescription and had limited adoption. The high-cost prescription treatment strategy reduces the probability of early treatment due to access and cost barriers, and eliminates complementary and synergistic benefits seen with many low-cost treatments.
|
viral culture+, 103.4% higher, RR 2.03, p = 0.33, treatment 6 of 117 (5.1%), control 3 of 119 (2.5%), days 6-20.
|
|
viral culture+, 31.1% lower, RR 0.69, p = 0.11, treatment 26 of 249 (10.4%), control 52 of 343 (15.2%), NNT 21, days 2-5.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
23.
Shah et al., SARS-CoV-2 infectious shedding and rebound among adults with and without oral antiviral use: two case-ascertained prospective household studies, The Lancet Microbe, doi:10.1016/j.lanmic.2025.101227.
Standing et al., 23 Feb 2024, Randomized Controlled Trial, United Kingdom, peer-reviewed, 54 authors, study period March 2022 - April 2022, trial ISRCTN30448031.
Contact: j.standing@ucl.ac.uk.
Abstract: Article
https://doi.org/10.1038/s41467-024-45641-0
Randomized controlled trial of molnupiravir
SARS-CoV-2 viral and antibody response in
at-risk adult outpatients
Received: 4 August 2023
A list of authors and their affiliations appears at the end of the paper
1234567890():,;
1234567890():,;
Accepted: 26 January 2024
Check for updates
Viral clearance, antibody response and the mutagenic effect of molnupiravir
has not been elucidated in at-risk populations. Non-hospitalised participants
within 5 days of SARS-CoV-2 symptoms randomised to receive molnupiravir
(n = 253) or Usual Care (n = 324) were recruited to study viral and antibody
dynamics and the effect of molnupiravir on viral whole genome sequence from
1437 viral genomes. Molnupiravir accelerates viral load decline, but virus is
detectable by Day 5 in most cases. At Day 14 (9 days post-treatment), molnupiravir is associated with significantly higher viral persistence and significantly
lower anti-SARS-CoV-2 spike antibody titres compared to Usual Care. Serial
sequencing reveals increased mutagenesis with molnupiravir treatment. Persistence of detectable viral RNA at Day 14 in the molnupiravir group is associated with higher transition mutations following treatment cessation. Viral
viability at Day 14 is similar in both groups with post-molnupiravir treated
samples cultured up to 9 days post cessation of treatment. The current 5-day
molnupiravir course is too short. Longer courses should be tested to reduce
the risk of potentially transmissible molnupiravir-mutated variants being
generated. Trial registration: ISRCTN30448031
Treatment of SARS-CoV-2 with the nucleoside analogue molnupiravir
(MK4482, EIDD2801) was reported to reduce viral load, hospitalisation
and mortality in unvaccinated participants with early COVID-19 in the
MOVeOUT trial1,2. Based on these data, molnupiravir received emergency use authorisation in the UK in November 2021 for early treatment of SARS-CoV-2 in individuals deemed to be at higher risk of
complications due to age or underlying comorbidities.
Molnupiravir is metabolised intracellularly to NHC-triphosphate,
which competes with natural cytidine and uridine for incorporation by
the viral RNA-dependent RNA polymerase (RdRp) into the nascent viral
RNA. This leads to abnormal, non-Watson-Crick pairing with guanosine
and uridine in further rounds of replication, increasing the substitution
of adenosine for guanosine and cytosine for uridine, so-called transition mutations, within the SARS-CoV-2 genome. Lethal mutagenesis
resulting from treatment with RdRp inhibitors eventually leads to viral
extinction3,4. A distinctive pattern of transition mutagenesis is evident
in viral genomes recovered from animals and humans who have
received molnupiravir3–5. The risk that, following molnupiravir treatment, some highly mutated viruses might remain viable and capable of
onward transmission has been postulated6,7.
To measure the impact of molnupiravir in a largely vaccinated
population, the Platform Adaptive trial of NOvel antiviRals for eArly
treatMent of covid-19 In the Community (PANORAMIC) was established. The first drug tested in PANORAMIC was molnupiravir, and
amongst 25,783 mostly vaccinated individuals, found that molnupiravir did not reduce hospitalisation or death8 (primary endpoint). Secondary outcomes showed those receiving molnupiravir experienced
significantly reduced viral load during treatment and reported faster
symptom recovery and fewer general..
DOI record:
{
"DOI": "10.1038/s41467-024-45641-0",
"ISSN": [
"2041-1723"
],
"URL": "http://dx.doi.org/10.1038/s41467-024-45641-0",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Viral clearance, antibody response and the mutagenic effect of molnupiravir has not been elucidated in at-risk populations. Non-hospitalised participants within 5 days of SARS-CoV-2 symptoms randomised to receive molnupiravir (n = 253) or Usual Care (n = 324) were recruited to study viral and antibody dynamics and the effect of molnupiravir on viral whole genome sequence from 1437 viral genomes. Molnupiravir accelerates viral load decline, but virus is detectable by Day 5 in most cases. At Day 14 (9 days post-treatment), molnupiravir is associated with significantly higher viral persistence and significantly lower anti-SARS-CoV-2 spike antibody titres compared to Usual Care. Serial sequencing reveals increased mutagenesis with molnupiravir treatment. Persistence of detectable viral RNA at Day 14 in the molnupiravir group is associated with higher transition mutations following treatment cessation. Viral viability at Day 14 is similar in both groups with post-molnupiravir treated samples cultured up to 9 days post cessation of treatment. The current 5-day molnupiravir course is too short. Longer courses should be tested to reduce the risk of potentially transmissible molnupiravir-mutated variants being generated. Trial registration: ISRCTN30448031</jats:p>",
"alternative-id": [
"45641"
],
"article-number": "1652",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "4 August 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "26 January 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "23 February 2024"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "J.F.S. has participated on a data safety monitoring board for GlaxoSmithKline (Sotrovimab) with fees paid to his institution. JSN-V-T was seconded to the Department of Health and Social Care, England (DHSC) from October 2017–March 2022. The views and opinions expressed in this paper are not necessarily those of DHSC or any of its arms-length bodies. JSN-V-T performed one-off paid consultancy for Merck Sharp and Dohme in June 2023, unrelated to the subject of the manuscript. K.H. was a member of the Health Technology Assessment General Committee and Funding Strategy Group until November 2022, and Research Professors Funding Committee at the UK National Institute for Health and Care Research (NIHR), received a grant from AstraZeneca (paid to their institution) to support a trial of Evusheld for the prevention of COVID-19 in high-risk individuals (RAPID-Protection), and was an independent member of the independent data monitoring committee for the OCTAVE-DUO trial of vaccines in individuals at high risk of COVID-19. D.M.L. has received grants or contracts from LifeArc, the UK Medical Research Council, Bristol Myers Squibb, GlaxoSmithKline, the British Society for Antimicrobial Chemotherapy, and Blood Cancer UK, personal fees or honoraria from Biotest UK, Gilead, and Merck, consulting fees from GlaxoSmithKline (paid to their institution), and conference support from Octapharma. DBR has received consulting fees from OMASS Therapeutics, GSK, and Sosei-Heptares and has a leadership and fiduciary role in the Heal-COVID trial TMG. M.L. is a member of the data monitoring and ethics committee of RAPIS-TEST (NIHR efficacy and mechanism evaluation). S.K. reports grants from GlaxoSmithKline, ViiV, Ridgeback Biotherapeutics, Vir, Merck, the UK Medical Research Council, and the Wellcome Trust (all paid to his institution), speaker’s honoraria from ViiV, and donations of drugs for clinical studies from ViiV Healthcare, Toyama, and GlaxoSmithKline. M.A. has received grants from the Blood and Transplant Research Unit, Janssen, Pfizer, Prenetics, Dunhill Medical Trust, the BMA Trust (Kathleen Harper Fund), and Antibiotic Research UK (all of which were paid to their institution), and consultancy fees from Prenetics and OxDx. M.A. reports a planned patent for Ramanomics, has participated on data safety monitoring boards or advisory boards for Prenetics, and has an unpaid leadership or fiduciary role in the E3 Initiative. NPBT has received payment for participation on an advisory board from MSD (before any knowledge or planning of this trial). O.v.H. has received consulting fees from MindGap (fees paid to Oxford University lnnovation), has participated on data safety monitoring boards or advisory boards for the CHICO trial, and has an unpaid leadership or fiduciary role in the British Society of Antimicrobial Chemotherapy. J.B. has received consulting fees from GlaxoSmithKline (paid to her institution). All other authors declare no competing interests."
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-4561-7173",
"affiliation": [],
"authenticated-orcid": false,
"family": "Standing",
"given": "Joseph F.",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-7404-7800",
"affiliation": [],
"authenticated-orcid": false,
"family": "Buggiotti",
"given": "Laura",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Guerra-Assuncao",
"given": "Jose Afonso",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Woodall",
"given": "Maximillian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ellis",
"given": "Samuel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Agyeman",
"given": "Akosua A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Miller",
"given": "Charles",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Okechukwu",
"given": "Mercy",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kirkpatrick",
"given": "Emily",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jacobs",
"given": "Amy I.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Williams",
"given": "Charlotte A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Roy",
"given": "Sunando",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Martin-Bernal",
"given": "Luz M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Williams",
"given": "Rachel",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8913-0009",
"affiliation": [],
"authenticated-orcid": false,
"family": "Smith",
"given": "Claire M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4177-2851",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sanderson",
"given": "Theo",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2946-3864",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ashford",
"given": "Fiona B.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Emmanuel",
"given": "Beena",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Afzal",
"given": "Zaheer M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5345-2156",
"affiliation": [],
"authenticated-orcid": false,
"family": "Shields",
"given": "Adrian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Richter",
"given": "Alex G.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6072-1430",
"affiliation": [],
"authenticated-orcid": false,
"family": "Dorward",
"given": "Jienchi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gbinigie",
"given": "Oghenekome",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Van Hecke",
"given": "Oliver",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lown",
"given": "Mark",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Francis",
"given": "Nick",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jani",
"given": "Bhautesh",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8093-7084",
"affiliation": [],
"authenticated-orcid": false,
"family": "Richards",
"given": "Duncan B.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1195-1680",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rahman",
"given": "Najib M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0331-7364",
"affiliation": [],
"authenticated-orcid": false,
"family": "Yu",
"given": "Ly-Mee",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Thomas",
"given": "Nicholas P. B.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8168-1746",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hart",
"given": "Nigel D.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5277-3545",
"affiliation": [],
"authenticated-orcid": false,
"family": "Evans",
"given": "Philip",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0619-1074",
"affiliation": [],
"authenticated-orcid": false,
"family": "Andersson",
"given": "Monique",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hayward",
"given": "Gail",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5268-8631",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hood",
"given": "Kerenza",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2579-4655",
"affiliation": [],
"authenticated-orcid": false,
"family": "Nguyen-Van-Tam",
"given": "Jonathan S.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Little",
"given": "Paul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hobbs",
"given": "F. D. Richard",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2769-0967",
"affiliation": [],
"authenticated-orcid": false,
"family": "Khoo",
"given": "Saye",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0102-3453",
"affiliation": [],
"authenticated-orcid": false,
"family": "Butler",
"given": "Christopher",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6102-2375",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lowe",
"given": "David M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8246-0534",
"affiliation": [],
"authenticated-orcid": false,
"family": "Breuer",
"given": "Judith",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Allen",
"given": "Julie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bayzid",
"given": "Nadua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Brown",
"given": "Julianne",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Burns",
"given": "Doug",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hadley",
"given": "Elizabeth",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hatcher",
"given": "Jim",
"sequence": "additional"
},
{
"affiliation": [],
"family": "McHugh",
"given": "Tim",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Thalasselis",
"given": "Chris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tomlinson",
"given": "Mia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yongblah",
"given": "Francis",
"sequence": "additional"
},
{
"affiliation": [],
"name": "PANORAMIC Virology Group",
"sequence": "additional"
}
],
"container-title": "Nature Communications",
"container-title-short": "Nat Commun",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
2,
23
]
],
"date-time": "2024-02-23T09:02:33Z",
"timestamp": 1708678953000
},
"deposited": {
"date-parts": [
[
2024,
2,
23
]
],
"date-time": "2024-02-23T09:03:09Z",
"timestamp": 1708678989000
},
"funder": [
{
"DOI": "10.13039/501100000272",
"award": [
"NIHR135366"
],
"doi-asserted-by": "publisher",
"name": "DH | National Institute for Health Research"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MR/X004724/1"
],
"doi-asserted-by": "publisher",
"name": "RCUK | Medical Research Council"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
24
]
],
"date-time": "2024-02-24T00:37:28Z",
"timestamp": 1708735048852
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
2,
23
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
2,
23
]
],
"date-time": "2024-02-23T00:00:00Z",
"timestamp": 1708646400000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
2,
23
]
],
"date-time": "2024-02-23T00:00:00Z",
"timestamp": 1708646400000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41467-024-45641-0.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-024-45641-0",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-024-45641-0.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2024,
2,
23
]
]
},
"published-online": {
"date-parts": [
[
2024,
2,
23
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1056/NEJMoa2116044",
"author": "A Jayk Bernal",
"doi-asserted-by": "publisher",
"first-page": "509",
"journal-title": "N. Engl. J. Med.",
"key": "45641_CR1",
"unstructured": "Jayk Bernal, A. et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 386, 509–520 (2022).",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1126/scitranslmed.abl7430",
"author": "WA Fischer",
"doi-asserted-by": "publisher",
"first-page": "eabl7430",
"journal-title": "Sci. Transl. Med",
"key": "45641_CR2",
"unstructured": "Fischer, W. A. et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med 14, eabl7430 (2022).",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1016/j.jbc.2021.100770",
"author": "CJ Gordon",
"doi-asserted-by": "publisher",
"first-page": "100770",
"journal-title": "template J. Biol. Chem.",
"key": "45641_CR3",
"unstructured": "Gordon, C. J., Tchesnokov, E. P., Schinazi, R. F. & Götte, M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA. template J. Biol. Chem. 297, 100770 (2021).",
"volume": "297",
"year": "2021"
},
{
"DOI": "10.1038/s41594-021-00651-0",
"author": "F Kabinger",
"doi-asserted-by": "publisher",
"first-page": "740",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "45641_CR4",
"unstructured": "Kabinger, F. et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 28, 740–746 (2021).",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1093/ve/veae001",
"doi-asserted-by": "publisher",
"key": "45641_CR5",
"unstructured": "Illingworth, C. J. R. et al. Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection. Virus Evol. veae001. https://doi.org/10.1093/ve/veae001 (2024)."
},
{
"key": "45641_CR6",
"unstructured": "November 30, 2021: Antimicrobial Drugs Advisory Committee Meeting Announcement—11/30/2021. FDA https://www.fda.gov/advisory-committees/advisory-committee-calendar/november-30-2021-antimicrobial-drugs-advisory-committee-meeting-announcement-11302021 (2022)."
},
{
"DOI": "10.1038/s41579-023-00878-2",
"author": "PV Markov",
"doi-asserted-by": "publisher",
"first-page": "361",
"journal-title": "Nat. Rev. Microbiol.",
"key": "45641_CR7",
"unstructured": "Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 21, 361–379 (2023).",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/S0140-6736(22)02597-1",
"author": "CC Butler",
"doi-asserted-by": "publisher",
"first-page": "281",
"journal-title": "Lancet",
"key": "45641_CR8",
"unstructured": "Butler, C. C. et al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet 401, 281–293 (2023).",
"volume": "401",
"year": "2023"
},
{
"DOI": "10.1038/s41467-022-34839-9",
"author": "I Donovan-Banfield",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "45641_CR9",
"unstructured": "Donovan-Banfield, I. et al. Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial. Nat. Commun. 13, 7284 (2022).",
"volume": "13",
"year": "2022"
},
{
"key": "45641_CR10",
"unstructured": "Pokay. https://github.com/nodrogluap/pokay (2023)."
},
{
"key": "45641_CR11",
"unstructured": "SARS2-ResistanceDB. https://github.com/ucl-pathgenomics/SARS2-ResistanceDB (2023)."
},
{
"key": "45641_CR12",
"unstructured": "Sanderson, T. Systematic Errors Associated with Some Implementations of ARTIC V4 and a Fast Workflow to Prescreen Samples for New Problematic Sites. https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473/16 (2021)."
},
{
"DOI": "10.1007/s15010-022-01959-9",
"doi-asserted-by": "publisher",
"key": "45641_CR13",
"unstructured": "Johnson, M. G. et al. Molnupiravir for the treatment of COVID-19 in immunocompromised participants: efficacy, safety, and virology results from the phase 3 randomized, placebo-controlled MOVe-OUT trial. Infection (2023) https://doi.org/10.1007/s15010-022-01959-9."
},
{
"DOI": "10.1371/journal.pmed.1004120",
"author": "DM Lowe",
"doi-asserted-by": "publisher",
"first-page": "e1004120",
"journal-title": "PLoS Med.",
"key": "45641_CR14",
"unstructured": "Lowe, D. M. et al. Favipiravir, lopinavir-ritonavir, or combination therapy (FLARE): A randomised, double-blind, 2 × 2 factorial placebo-controlled trial of early antiviral therapy in COVID-19. PLoS Med. 19, e1004120 (2022).",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.1016/S2666-5247(23)00005-8",
"author": "S Galmiche",
"doi-asserted-by": "publisher",
"first-page": "e409",
"journal-title": "Lancet Microbe",
"key": "45641_CR15",
"unstructured": "Galmiche, S. et al. SARS-CoV-2 incubation period across variants of concern, individual factors, and circumstances of infection in France: a case series analysis from the ComCor study. Lancet Microbe 4, e409–e417 (2023).",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.1002/psp4.12543",
"author": "A Gonçalves",
"doi-asserted-by": "publisher",
"first-page": "509",
"journal-title": "CPT Pharmacomet. Syst. Pharmacol.",
"key": "45641_CR16",
"unstructured": "Gonçalves, A. et al. Timing of antiviral treatment initiation is critical to reduce SARS-CoV-2 viral load. CPT Pharmacomet. Syst. Pharmacol. 9, 509–514 (2020).",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1002/cpt.2223",
"author": "S Gastine",
"doi-asserted-by": "publisher",
"first-page": "321",
"journal-title": "Clin. Pharmacol. Ther.",
"key": "45641_CR17",
"unstructured": "Gastine, S. et al. Systematic review and patient-level meta-analysis of SARS-CoV-2 viral dynamics to model response to antiviral therapies. Clin. Pharmacol. Ther. 110, 321–333 (2021).",
"volume": "110",
"year": "2021"
},
{
"DOI": "10.1001/jamapediatrics.2022.3261",
"author": "PS Walsh",
"doi-asserted-by": "publisher",
"first-page": "e223261",
"journal-title": "JAMA Pediatr.",
"key": "45641_CR18",
"unstructured": "Walsh, P. S. et al. Association of early oseltamivir with improved outcomes in hospitalized children with influenza, 2007–2020. JAMA Pediatr. 176, e223261 (2022).",
"volume": "176",
"year": "2022"
},
{
"DOI": "10.1016/S2666-5247(21)00305-0",
"author": "B Barin",
"doi-asserted-by": "publisher",
"first-page": "e274",
"journal-title": "Lancet Microbe",
"key": "45641_CR19",
"unstructured": "Barin, B., Kasap, U., Selçuk, F., Volkan, E. & Uluçkan, Ö. Comparison of SARS-CoV-2 anti-spike receptor binding domain IgG antibody responses after CoronaVac, BNT162b2, ChAdOx1 COVID-19 vaccines, and a single booster dose: a prospective, longitudinal population-based study. Lancet Microbe 3, e274–e283 (2022).",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1093/ofid/ofac618",
"author": "C Moser",
"doi-asserted-by": "publisher",
"first-page": "ofac618",
"journal-title": "Open Forum Infect. Dis.",
"key": "45641_CR20",
"unstructured": "Moser, C. et al. Predictors of SARS-CoV-2 RNA from nasopharyngeal swabs and concordance with other compartments in nonhospitalized adults with mild to moderate COVID-19. Open Forum Infect. Dis. 9, ofac618 (2022).",
"volume": "9",
"year": "2022"
},
{
"DOI": "10.1038/s41467-021-26479-2",
"author": "J Wei",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "45641_CR21",
"unstructured": "Wei, J. et al. Anti-spike antibody response to natural SARS-CoV-2 infection in the general population. Nat. Commun. 12, 6250 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41591-023-02414-4",
"doi-asserted-by": "publisher",
"key": "45641_CR22",
"unstructured": "Barnes, E. et al. SARS-CoV-2-specific immune responses and clinical outcomes after COVID-19 vaccination in patients with immune-suppressive disease. Nat. Med. https://doi.org/10.1038/s41591-023-02414-4 (2023)."
},
{
"DOI": "10.1093/ofid/ofad154",
"author": "AF Carlin",
"doi-asserted-by": "publisher",
"first-page": "ofad154",
"journal-title": "Open Forum Infect. Dis.",
"key": "45641_CR23",
"unstructured": "Carlin, A. F. et al. Neutralizing antibody responses after severe acute respiratory syndrome coronavirus 2 BA.2 and BA.2.12.1 infection do not neutralize BA.4 and BA.5 and can be blunted by nirmatrelvir/ritonavir treatment. Open Forum Infect. Dis. 10, ofad154 (2023).",
"volume": "10",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(22)00644-2",
"author": "SH Khoo",
"doi-asserted-by": "publisher",
"first-page": "183",
"journal-title": "Lancet Infect. Dis.",
"key": "45641_CR24",
"unstructured": "Khoo, S. H. et al. Molnupiravir versus placebo in unvaccinated and vaccinated patients with early SARS-CoV-2 infection in the UK (AGILE CST-2): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Infect. Dis. 23, 183–195 (2023).",
"volume": "23",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06649-6",
"doi-asserted-by": "publisher",
"key": "45641_CR25",
"unstructured": "Sanderson, T. et al. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature https://doi.org/10.1038/s41586-023-06649-6 (2023)."
},
{
"DOI": "10.1093/molbev/msq024",
"author": "JE Barrick",
"doi-asserted-by": "publisher",
"first-page": "1338",
"journal-title": "Mol. Biol. Evol.",
"key": "45641_CR26",
"unstructured": "Barrick, J. E., Kauth, M. R., Strelioff, C. C. & Lenski, R. E. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27, 1338–1347 (2010).",
"volume": "27",
"year": "2010"
},
{
"DOI": "10.1016/j.chom.2020.11.007",
"author": "AJ Greaney",
"doi-asserted-by": "publisher",
"first-page": "44",
"journal-title": "Cell Host Microbe",
"key": "45641_CR27",
"unstructured": "Greaney, A. J. et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9 (2021).",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.1101/2021.02.17.431683",
"doi-asserted-by": "crossref",
"key": "45641_CR28",
"unstructured": "Starr, T. N., Greaney, A. J., Dingens, A. S. & Bloom, J. D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2 (2021)."
},
{
"DOI": "10.1038/s41564-021-00954-4",
"author": "J Zahradník",
"doi-asserted-by": "publisher",
"first-page": "1188",
"journal-title": "Nat. Microbiol.",
"key": "45641_CR29",
"unstructured": "Zahradník, J. et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 6, 1188–1198 (2021).",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1101/2021.03.22.436481",
"doi-asserted-by": "publisher",
"key": "45641_CR30",
"unstructured": "Sun, Z. et al. Neutralization of European, South African, and United States SARS-CoV-2 mutants by a human antibody and antibody domains. Preprint at https://doi.org/10.1101/2021.03.22.436481 (2021)."
},
{
"DOI": "10.1016/j.isci.2023.107786",
"author": "A Zibat",
"doi-asserted-by": "publisher",
"first-page": "107786",
"journal-title": "iScience",
"key": "45641_CR31",
"unstructured": "Zibat, A. et al. N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody. iScience 26, 107786 (2023).",
"volume": "26",
"year": "2023"
},
{
"DOI": "10.1038/s41467-019-13940-6",
"author": "TP Sheahan",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "45641_CR32",
"unstructured": "Sheahan, T. P. et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 11, 222 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1002/jmv.27285",
"author": "FAT Boshier",
"doi-asserted-by": "publisher",
"first-page": "161",
"journal-title": "J. Med. Virol.",
"key": "45641_CR33",
"unstructured": "Boshier, F. A. T. et al. Evolution of viral variants in remdesivir-treated and untreated SARS-CoV-2-infected pediatrics patients. J. Med. Virol. 94, 161–172 (2022).",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1093/cid/ciaa023",
"author": "CK Lumby",
"doi-asserted-by": "publisher",
"first-page": "e191",
"journal-title": "Clin. Infect. Dis.",
"key": "45641_CR34",
"unstructured": "Lumby, C. K. et al. Favipiravir and Zanamivir Cleared Infection with Influenza B in a Severely Immunocompromised Child. Clin. Infect. Dis. 71, e191–e194 (2020).",
"volume": "71",
"year": "2020"
},
{
"DOI": "10.1016/j.xcrm.2020.100144",
"author": "AA Mueller",
"doi-asserted-by": "publisher",
"first-page": "100144",
"journal-title": "Cell Rep. Med.",
"key": "45641_CR35",
"unstructured": "Mueller, A. A. et al. Inflammatory biomarker trends predict respiratory decline in COVID-19 patients. Cell Rep. Med. 1, 100144 (2020).",
"volume": "1",
"year": "2020"
},
{
"DOI": "10.7326/M22-0729",
"author": "MG Johnson",
"doi-asserted-by": "publisher",
"first-page": "1126",
"journal-title": "Ann. Intern. Med.",
"key": "45641_CR36",
"unstructured": "Johnson, M. G. et al. Effect of molnupiravir on biomarkers, respiratory interventions, and medical services in COVID-19: a randomized, placebo-controlled trial. Ann. Intern. Med. 175, 1126–1134 (2022).",
"volume": "175",
"year": "2022"
},
{
"DOI": "10.1186/s13073-021-00839-5",
"author": "DJ Baker",
"doi-asserted-by": "publisher",
"journal-title": "Genome Med",
"key": "45641_CR37",
"unstructured": "Baker, D. J. et al. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med 13, 21 (2021).",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.3201/eid2612.203309",
"author": "GL Morley",
"doi-asserted-by": "publisher",
"first-page": "2970",
"journal-title": "Emerg. Infect. Dis.",
"key": "45641_CR38",
"unstructured": "Morley, G. L. et al. Sensitive detection of SARS-CoV-2-specific antibodies in dried blood spot samples. Emerg. Infect. Dis. 26, 2970–2973 (2020).",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1111/bcp.15518",
"author": "AA Agyeman",
"doi-asserted-by": "publisher",
"first-page": "5428",
"journal-title": "Br. J. Clin. Pharmacol.",
"key": "45641_CR39",
"unstructured": "Agyeman, A. A. et al. Comparative assessment of viral dynamic models for SARS-CoV-2 for pharmacodynamic assessment in early treatment trials. Br. J. Clin. Pharmacol. 88, 5428–5433 (2022).",
"volume": "88",
"year": "2022"
},
{
"DOI": "10.1023/A:1012299115260",
"author": "SL Beal",
"doi-asserted-by": "publisher",
"first-page": "481",
"journal-title": "J. Pharmacokinet. Pharmacodyn.",
"key": "45641_CR40",
"unstructured": "Beal, S. L. Ways to fit a PK model with some data below the quantification limit. J. Pharmacokinet. Pharmacodyn. 28, 481–504 (2001).",
"volume": "28",
"year": "2001"
},
{
"DOI": "10.5281/zenodo.7764938",
"doi-asserted-by": "publisher",
"key": "45641_CR41",
"unstructured": "Patel, H. et al. nf-core/viralrecon: nf-core/viralrecon v2.6.0 - Rhodium Raccoon (2023). https://doi.org/10.5281/zenodo.7764938."
},
{
"DOI": "10.1186/s13059-018-1618-7",
"author": "ND Grubaugh",
"doi-asserted-by": "publisher",
"journal-title": "Genome Biol.",
"key": "45641_CR42",
"unstructured": "Grubaugh, N. D. et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 20, 8 (2019).",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1128/JVI.03181-13",
"author": "I Monne",
"doi-asserted-by": "publisher",
"first-page": "4375",
"journal-title": "J. Virol.",
"key": "45641_CR43",
"unstructured": "Monne, I. et al. Emergence of a highly pathogenic avian influenza virus from a low-pathogenic progenitor. J. Virol. 88, 4375–4388 (2014).",
"volume": "88",
"year": "2014"
},
{
"DOI": "10.1093/molbev/mst010",
"author": "K Katoh",
"doi-asserted-by": "publisher",
"first-page": "772",
"journal-title": "Mol. Biol. Evol.",
"key": "45641_CR44",
"unstructured": "Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).",
"volume": "30",
"year": "2013"
},
{
"DOI": "10.1093/molbev/msu300",
"author": "L-T Nguyen",
"doi-asserted-by": "publisher",
"first-page": "268",
"journal-title": "Mol. Biol. Evol.",
"key": "45641_CR45",
"unstructured": "Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).",
"volume": "32",
"year": "2015"
},
{
"DOI": "10.1038/s41588-021-00862-7",
"author": "Y Turakhia",
"doi-asserted-by": "publisher",
"first-page": "809",
"journal-title": "Nat. Genet.",
"key": "45641_CR46",
"unstructured": "Turakhia, Y. et al. Ultrafast sample placement on existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat. Genet. 53, 809–816 (2021).",
"volume": "53",
"year": "2021"
}
],
"reference-count": 46,
"references-count": 46,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41467-024-45641-0"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Physics and Astronomy",
"General Biochemistry, Genetics and Molecular Biology",
"General Chemistry",
"Multidisciplinary"
],
"subtitle": [],
"title": "Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "15"
}