The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country
et al., Intervirology, doi:10.1159/000540282, Nov 2024
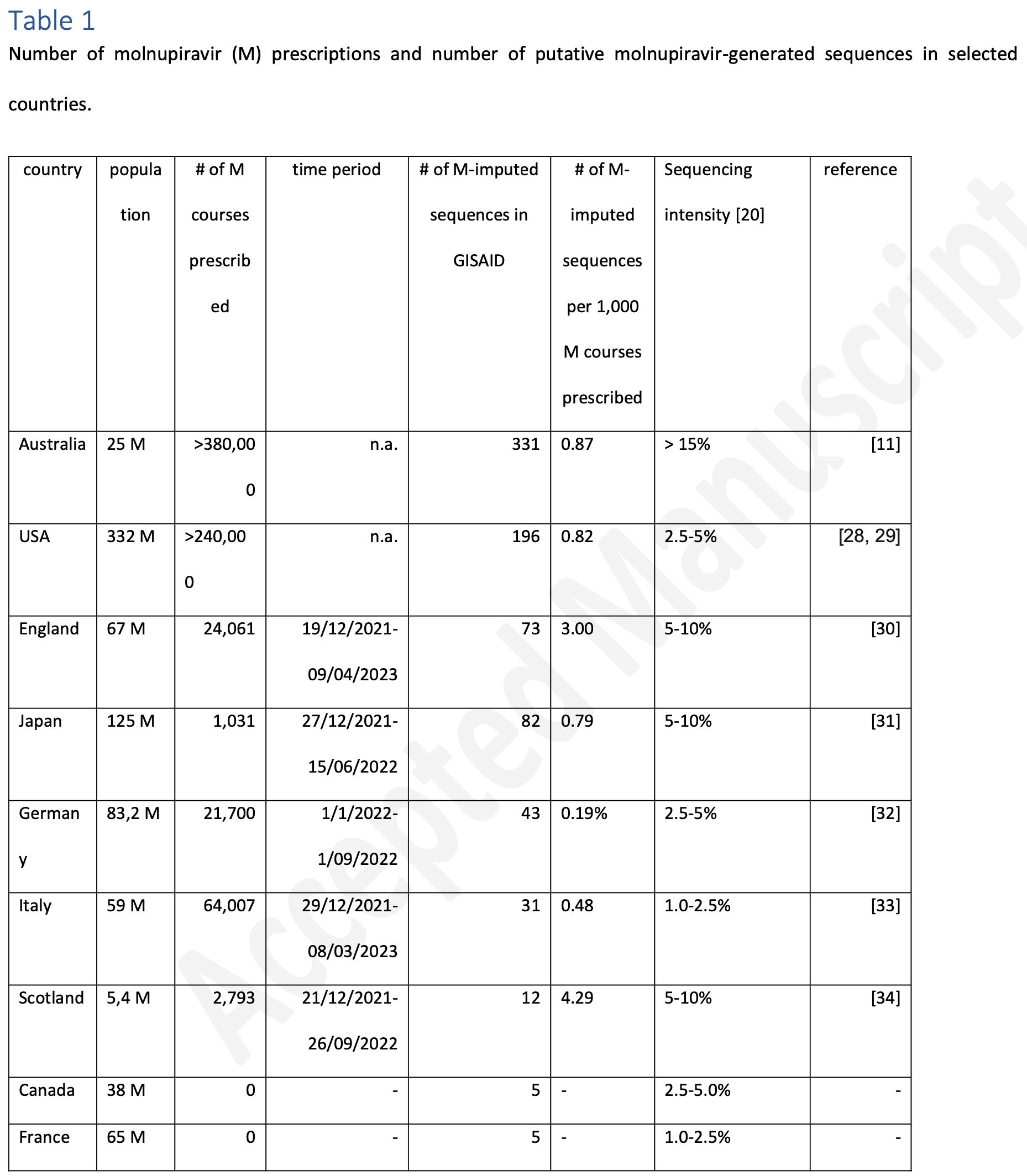
Analysis of GISAID SARS-CoV-2 sequences showing a strong correlation between molnupiravir prescriptions and putative molnupiravir-generated variants by country. Authors identify 1,094 sequences with a molnupiravir mutational signature, predominantly from countries authorizing widespread use of the drug. The results suggest molnupiravir can generate fit SARS-CoV-2 variants that are able to transmit in the general population.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Focosi et al., 8 Nov 2024, Italy, peer-reviewed, 3 authors.
Contact: daniele.focosi@gmail.com.
The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country.
Intervirology, doi:10.1159/000540282
Introduction : Molnupiravir is one of the oral direct-acting antivirals against SARS-CoV-2, largely deployed during the COVID-19 pandemic since the 2022 Omicron wave. While efficacy has been questioned in post-marketing clinical trials (leading to the EMA withdrawing its authorization), growing concerns have mounted regarding its possible mutagenic effects on the virus. While it has been assumed that either all the host viral load was cleared by the drug or that drug-generated variants were not fit enough to survive, several lineages with a high transition/transversion ratio (a signature of molnupiravir action) have been recently reported from GISAID. Methods : We report here a systematic analysis of the GISAID database for sequences showing a molnupiravir signature, exposing a public web-based interface ( https://ukcovid.xyz/molnupiravir/ ), and performing an imputation analysis based on per-country prescription (corrected by sequencing). Results: Our analysis confirms a direct correlation between the number of molnupiravir courses and the number of mutationally signed deposited in GISAID in individual countries. Conclusions: Molnupiravir can generate fit SARS-CoV-2 variants that transmit in the general population.
Ethics statement: An ethics statement is not applicable because this study is based exclusively on published literature. Author contributions: D.M. analyzed the GISAID database, created the public web repository, and revised the manuscript; D.F. retrieved prescription counts by country and wrote the first draft; F.M. revised the manuscript. Conflict of interest disclosure: We declare that we have no conflict of interests related to this manuscript.
References
Alteri, Fox, Scutari, Burastero, Volpi et al., A proof-of-concept study on the genomic evolution of Sars-Cov-2 in molnupiravir-treated, paxlovid-treated and drug-naïve patients, Commun Biol
Alteri, Fox, Scutari, Burastero, Volpi et al., A proof-of-concept study on the genomic evolution of Sars-Cov-2 in molnupiravir-treated, paxlovid-treated and drug-naïve patients, Commun Biol
Bernal, Da Silva, Musungaie, Kovalchuk, Gonzalez et al., Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients, N Engl J Med
Bloom, Beichman, Neher, Harris, Evolution of the SARS-CoV-2 mutational spectrum, Molecular biology and evolution
Chen, Azman, Chen, Zou, Tian et al., Global landscape of SARS-CoV-2 genomic surveillance and data sharing, Nature genetics
Donovan-Banfield, Penrice-Randal, Goldswain, Rzeszutek, Pilgrim et al., Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial, Nat Commun
El-Haddad, Adhikari, Tu, Cheng, Leng et al., Intra-host mutation rate of acute SARS-CoV-2 infection during the initial pandemic wave, Virus genes
Fischer, Eron, Holman, Cohen, Fang et al., A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus
Fletcher, Ia, Penrice-Randal, Characterisation of SARS-CoV-2 genomic variations in response to molnupiravir treatment in the AGILE Phase IIa clinical trial, doi:10.21203/rs.3.rs-1835695/v1
Focosi, Molnupiravir: From Hope to Epic Fail
Fountain-Jones, Vanhaeften, Williamson, Maskell, Chua et al., Antiviral treatments lead to the rapid accrual of hundreds of SARS-CoV-2 mutations in immunocompromised patients
Gold, Kelleher, Magid, Jackson, Pennini et al., Dispensing of Oral Antiviral Drugs for Treatment of COVID-19 by Zip Code-Level Social Vulnerability -United States, December 23, 2021
Hisner, LongDesertTrain) tweet
Illingworth, Guerra-Assuncao, Gregg, Charles, Pang et al., Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection
Khare, Gurry, Freitas, Schultz, Bach et al., GISAID's Role in Pandemic Response, China CDC Wkly
Kimata, Watanabe, Yanagida, Kinoshita, Maekawa, Safety and Effectiveness of Molnupiravir (LAGEVRIO(®)) Capsules in Japanese Patients with COVID-19: Interim Report of Post-marketing Surveillance in Japan, Infectious diseases and therapy
Neher, SARS-CoV-2 variant rates
Penrice-Randal, Bentley, Sharma, Kirby, Ia et al., The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in infected and immune suppressed mice, bioRxiv
Plata, The black market for covid-19 antiviral drugs
Robert F Service, A prominent virologist warns COVID-19 pill could unleash dangerous mutants. Others see little cause for alarm
Sanderson, Hisner, Ia, Peacock, Ruis, A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature
Sheahan, Sims, Zhou, Graham, Pruijssers et al., An orally bioavailable broadspectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Science translational medicine
Sheahan, Stevens, Narowski, Krajewski, Lee et al., The Antiviral Mechanism of Action of Molnupiravir in Humans with COVID-19, medRxiv
Sonnleitner, Prelog, Sonnleitner, Hinterbichler, Halbfurter et al., Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host, Nat Commun
Sublineage, 2 with 8 additional spike mutations (9 seq, Australia), PANGO
Swanstrom, Schinazi, Lethal mutagenesis as an antiviral strategy, Science
Tibble, Mueller, Proud, Hall, Kurdi et al., Uptake of monoclonal antibodies and antiviral therapies for COVID-19 in Scotland, Lancet
William, Haseltine, Supercharging New Viral Variants: The Dangers Of Molnupiravir (Part 1)
DOI record:
{
"DOI": "10.1159/000540282",
"ISSN": [
"0300-5526",
"1423-0100"
],
"URL": "http://dx.doi.org/10.1159/000540282",
"abstract": "<jats:p>Introduction : Molnupiravir is one of the oral direct-acting antivirals against SARS-CoV-2, largely deployed during the COVID-19 pandemic since the 2022 Omicron wave. While efficacy has been questioned in post-marketing clinical trials (leading to the EMA withdrawing its authorization), growing concerns have mounted regarding its possible mutagenic effects on the virus. While it has been assumed that either all the host viral load was cleared by the drug or that drug-generated variants were not fit enough to survive, several lineages with a high transition/transversion ratio (a signature of molnupiravir action) have been recently reported from GISAID. \nMethods : We report here a systematic analysis of the GISAID database for sequences showing a molnupiravir signature, exposing a public web-based interface (https://ukcovid.xyz/molnupiravir/ ), and performing an imputation analysis based on per-country prescription (corrected by sequencing). \nResults: Our analysis confirms a direct correlation between the number of molnupiravir courses and the number of mutationally signed deposited in GISAID in individual countries. \nConclusions: Molnupiravir can generate fit SARS-CoV-2 variants that transmit in the general population.\n</jats:p>",
"author": [
{
"affiliation": [],
"family": "Focosi",
"given": "Daniele",
"sequence": "first"
},
{
"affiliation": [],
"family": "McNally",
"given": "Dave",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Maggi",
"given": "Fabrizio",
"sequence": "additional"
}
],
"container-title": "Intervirology",
"container-title-short": "Intervirology",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
11,
9
]
],
"date-time": "2024-11-09T17:00:23Z",
"timestamp": 1731171623000
},
"deposited": {
"date-parts": [
[
2024,
11,
9
]
],
"date-time": "2024-11-09T17:00:25Z",
"timestamp": 1731171625000
},
"indexed": {
"date-parts": [
[
2024,
11,
9
]
],
"date-time": "2024-11-09T17:40:09Z",
"timestamp": 1731174009614,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
11,
8
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
8
]
],
"date-time": "2024-11-08T00:00:00Z",
"timestamp": 1731024000000
}
},
{
"URL": "https://creativecommons.org/licenses/by-nc/4.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
8
]
],
"date-time": "2024-11-08T00:00:00Z",
"timestamp": 1731024000000
}
}
],
"link": [
{
"URL": "https://karger.com/int/article-pdf/doi/10.1159/000540282/4298792/000540282.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://karger.com/int/article-pdf/doi/10.1159/000540282/4298792/000540282.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "127",
"original-title": [],
"page": "1-9",
"prefix": "10.1159",
"published": {
"date-parts": [
[
2024,
11,
8
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
8
]
]
},
"publisher": "S. Karger AG",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://karger.com/doi/10.1159/000540282"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country.",
"type": "journal-article"
}