Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment
et al., J. Cellular Physiology, doi:10.1002/jcp.30055, Sep 2020
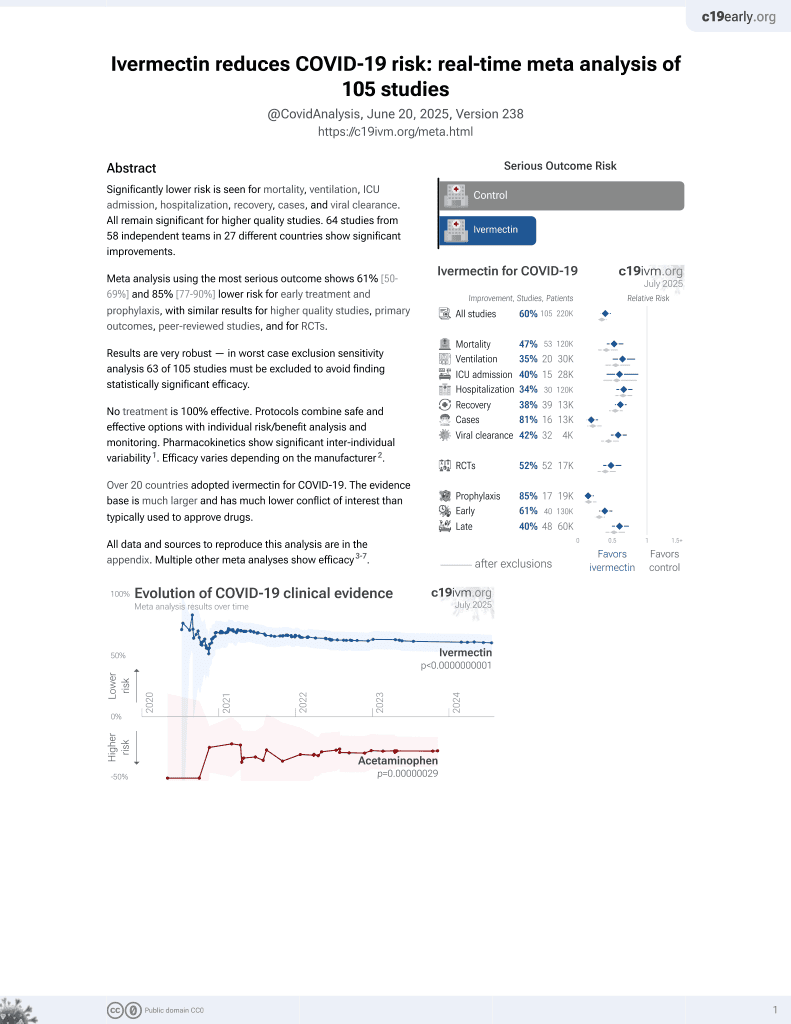
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study showing Ivermectin is a safe wide-spectrum antiviral against SARS-CoV-2, human papillomavirus (HPV), Epstein-Barr virus (EBV), and HIV.
Authors note that the combination of ivermectin and other drugs might result in more favorable prognoses for patients with COVID-19, for example ivermerctin and HCQ.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Li et al., 22 Sep 2020, peer-reviewed, 3 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Quantitative proteomics reveals a broad‐spectrum antiviral property of ivermectin, benefiting for COVID‐19 treatment
Journal of Cellular Physiology, doi:10.1002/jcp.30055
Viruses such as human cytomegalovirus (HCMV), human papillomavirus (HPV), Epstein-Barr virus (EBV), human immunodeficiency virus (HIV), and coronavirus (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) represent a great burden to human health worldwide. FDA-approved anti-parasite drug ivermectin is also an antibacterial, antiviral, and anticancer agent, which offers more potentiality to improve global public health, and it can effectively inhibit the replication of SARS-CoV-2 in vitro. This study sought to identify ivermectin-related virus infection pathway alterations in human ovarian cancer cells. Stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics was used to analyze human ovarian cancer cells TOV-21G treated with and without ivermectin (20 μmol/L) for 24 h, which identified 4447 ivermectin-related proteins in ovarian cancer cells. Pathway network analysis revealed four statistically significant antiviral pathways, including HCMV, HPV, EBV, and HIV1 infection pathways. Interestingly, compared with the reported 284 SARS-CoV-2/COVID-19-related genes from GencLip3, we identified 52 SARS-CoV-2/COVID-19-related protein alterations when treated with and without ivermectin. Protein-protein network (PPI) was constructed based on the interactions between 284 SARS-CoV-2/COVID-19-related genes and between 52 SARS-CoV-2/COVID-19-related proteins regulated by ivermectin. Molecular complex detection analysis of PPI network identified three hub modules, including cytokines and growth factor family, MAP kinase and G-protein family, and HLA class proteins. Gene Ontology analysis revealed 10 statistically significant cellular components, 13 molecular functions, and 11 biological processes. These findings demonstrate the broad-spectrum antiviral property of ivermectin benefiting for COVID-19 treatment in the context of predictive, preventive, and personalized medicine in virus-related diseases.
CONFLICT OF INTERESTS The authors have declared that no competing interests exist.
AUTHOR CONTRIBUTIONS Na Li performed SILAC cell experiments, analyzed the data, prepared figures and tables, and drafted the manuscript. Lingfeng Zhao participated in bioinformatics analysis. Xianquan Zhan conceived the concept, guided experiments and data analysis, supervised results, wrote and critically revised the manuscript, and was responsible for the financial supports and corresponding works. All authors approved the final manuscript.
ORCID
Xianquan Zhan http://orcid.org/0000-0002-4984-3549
SUPPORTING INFORMATION Additional Supporting Information may be found online in the supporting information tab for this article. How to cite this article: Li N, Zhao L, Zhan X. Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment. J Cell Physiol. 2021;236:2959-2975. https://doi.org/10.1002/jcp.30055
References
Abdeltawab, Rifaie, Shoeib, El-Latif, Badawi et al., Insights into the impact of Ivermectin on some protein aspects linked to Culex pipiens digestion and immunity, Parasitology Research
Almeida, Queiroz, Sousa, Sousa, Cervical cancer and HPV infection: Ongoing therapeutic research to counteract the action of E6 and E7 oncoproteins, Drug Discovery Today
Alpalhão, Ferreira, Filipe, Persistent SARS-CoV-2 infection and the risk for cancer, Medical Hypotheses
Andoniou, Degli-Esposti, Insights into the mechanisms of CMV-mediated interference with cellular apoptosis, Immunology and Cell Biology
Ashour, Ivermectin: From theory to clinical application, International Journal of Antimicrobial Agents
Athanasiou, Bowden, Paraskevaidi, Fotopoulou, Martin-Hirsch et al., HPV vaccination and cancer prevention, Best Practice & Research Clinical Obstetrics & Gynaecology
Bader, Hogue, An automated method for finding molecular complexes in large protein interaction networks, BMC Bioinformatics
Benvenuto, Giovanetti, Ciccozzi, Spoto, Angeletti et al., The 2019-new coronavirus epidemic: Evidence for virus evolution, Journal of Medical Virology
Bindea, Mlecnik, Hackl, Charoentong, Tosolini et al., ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks, Bioinformatics
Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature
Boussinesq, Ivermectin, Medecine tropicale
Britt, Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease, Current Topics in Microbiology and Immunology
Buechner, Common skin disorders of the penis, BJU International
Burg, Miller, Baker, Birnbaum, Currie et al., Avermectins, new family of potent anthelmintic agents: Producing organism and fermentation, Antimicrobial Agents Chemother
Buxmann, Hamprecht, Meyer-Wittkopf, Friese, Primary human cytomegalovirus (HCMV) infection in pregnancy, Deutsches Ärzteblatt International
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research
Chabala, Mrozik, Tolman, Eskola, Lusi et al., Ivermectin, a new broad-spectrum antiparasitic agent, Journal of Medicinal Chemistry
Chaccour, Hammann, Ramón-García, Rabinovich, Ivermectin and COVID-19: Keeping rigor in times of urgency, American Journal of Tropical Medicine and Hygiene
Crump, Ivermectin: Enigmatic multifaceted 'wonder' drug continues to surprise and exceed expectations, Journal of Antibiotics
Csóka, Németh, Szabó, Davies, Varga et al., Macrophage P2X4 receptors augment bacterial killing and protect against sepsis, JCI Insight
Deng, Xu, Long, Xie, Suppressing ROS-TFE3-dependent autophagy enhances ivermectin-induced apoptosis in human melanoma cells, Journal of Cellular Biochemistry, doi:10.1002/jcb.27490
Develoux, None
Diao, Cheng, Zhao, Xu, Dong et al., Ivermectin inhibits canine mammary tumor growth by LI ET AL. | 2973 regulating cell cycle progression and WNT signaling, BMC Veterinary Research
Dou, Chen, Wang, Yuan, Lei et al., Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer, Cancer Research
Gallardo, Mariamé, Gence, Tilkin-Mariamé, Macrocyclic lactones inhibit nasopharyngeal carcinoma cells proliferation through PAK1 inhibition and reduce in vivo tumor growth, Drug Design, Development and Therapy
Guzzo, Furtek, Porras, Chen, Tipping et al., Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects, Journal of Clinical Pharmacology
He, Ding, Zhang, Che, He et al., Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS, Journal of Pathology
Holder, Grant, Human cytomegalovirus IL-10 augments NK cell cytotoxicity, Journal of Leukocyte Biology
Hoppe-Seyler, Bossler, Braun, Herrmann, Hoppe-Seyler, The HPV E6/E7 Oncogenes: Key factors for viral carcinogenesis and therapeutic targets, Trends in Microbiology
Huynh, Gulick, HIV prevention. Treasure Island
Jiang, Wang, Sun, Wu, Ivermectin reverses the drug resistance in cancer cells through EGFR/ERK/Akt/NF-κB pathway, Journal of Experimental and Clinical Cancer Research
Juarez, Schcolnik-Cabrera, Dueñas-Gonzalez, The multitargeted drug ivermectin: From an antiparasitic agent to a repositioned cancer drug, American Journal of Cancer Research
Lai, Shih, Ko, Tang, Hsueh, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges, International Journal of Antimicrobial Agents
Laing, Gillan, Devaney, Ivermectin-Old drug, new tricks?, Trends Parasitol
Lee, Lim, Ham, Kim, You et al., Ivermectin induces apoptosis of porcine trophectoderm and uterine luminal epithelial cells through loss of mitochondrial membrane potential, mitochondrial calcium ion overload, and reactive oxygen species generation, Pesticide Biochemistry and Physiology
Letko, Marzi, Munster, Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses, Nature Microbiology
Levy, Martin, Bursztejn, Chiaverini, Miquel et al., Ivermectin safety in infants and children under 15 kg treated for scabies: A multicentric observational study, British Journal of Dermatology
Li, Zhan, Anti-parasite drug ivermectin can suppress ovarian cancer by regulating lncRNA-EIF4A3-mRNA axes, EPMA Journal
Li, Zhou, Zhang, Wang, Zhao et al., Updated Approaches against SARS-CoV-2, Antimicrob Agents Chemother
Lv, Liu, Wang, Dang, Qiu et al., Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo, Antiviral Research
Marques-Piubelli, Salas, Pachas, Becker-Hecker, Vega et al., Epstein-Barr virus-associated B-cell lymphoproliferative disorders and lymphomas: A review, Pathology
Nash, Robertson, How to evolve the response to the global HIV epidemic with new metrics and targets based on pretreatment CD4 counts, Current HIV/AIDS Reports
Nicolas, Maia, Bassat, Kobylinski, Monteiro et al., Safety of oral ivermectin during pregnancy: A systematic review and meta-analysis, Lancet Global Health
Nussinovitch, Prais, Volovitz, Shapiro, Amir, Post-infectious acute cerebellar ataxia in children, Clinical Pediatrics
Patrì, Fabbrocini, Hydroxychloroquine and ivermectin: A synergistic combination for COVID-19 chemoprophylaxis and treatment?, Journal of the American Academy of Dermatology
Rappsilber, Ishihama, Mann, Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics, Analytical Chemistry
Rehwinkel, Gack, RIG-I-like receptors: Their regulation and roles in RNA sensing, Nature Reviews Immunology
Rezk, Zhao, Weiss, Epstein-Barr virus (EBV)-associated lymphoid proliferations, a 2018 update, Human Pathology
Schaller, Gonser, Belge, Braunsdorf, Nordin et al., Dual anti-inflammatory and anti-parasitic action of topical ivermectin 1% in papulopustular rosacea, Journal of the European Academy of Dermatology and Venereology
Shannon-Lowe, Rowe, Epstein Barr virus entry; kissing and conjugation, Current Opinion in Virology
Szklarczyk, Franceschini, Wyder, Forslund, Heller et al., STRING v10: Protein-protein interaction networks, integrated over the tree of life, Nucleic Acids Research
Ulrich, Pillat, CD147 as a target for COVID-19 treatment: Suggested effects of azithromycin and stem cell engagement, Stem Cell Reviews and Reports
Van Wyk, Malan, Resistance of field strains of Haemonchus contortus to ivermectin, closantel, rafoxanide and the benzimidazoles in South Africa, Veterinary Record
Varghese, Kaukinen, Gläsker, Bespalov, Hanski et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research
Wagstaff, Sivakumaran, Heaton, Harrich, Jans, Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal
Wang, Zhao, Wang, Wen, Jiang et al., GenCLiP 3: Mining human genes' functions and regulatory networks from PubMed based on co-occurrences and natural language processing, Bioinformatics
Wei, Zang, Li, Zhang, Gao et al., Grouper PKR activation inhibits red-spotted grouper nervous necrosis virus (RGNNV) replication in infected cells, Developmental and Comparative Immunology
Yang, Atkinson, Wang, Lee, Bogoyevitch et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research
Yu, Wang, Han, He, ClusterProfiler: An R package for comparing biological themes among gene clusters, OMICS
Zavattoni, Furione, Arossa, Iasci, Spinillo et al., Diagnosis and counseling of fetal and neonatal HCMV infection, Early Human Development
Zhang, Song, Xiong, Ci, Li et al., Inhibitory effects of ivermectin on nitric oxide and prostaglandin E2 production in LPS-stimulated RAW 264.7 macrophages, International Immunopharmacology
Zhang, Wang, Wang, Zheng, Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-κB activation by interacting with p65/RelA and p50/NF-κB1, Journal of Virology
DOI record:
{
"DOI": "10.1002/jcp.30055",
"ISSN": [
"0021-9541",
"1097-4652"
],
"URL": "http://dx.doi.org/10.1002/jcp.30055",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Viruses such as human cytomegalovirus (HCMV), human papillomavirus (HPV), Epstein–Barr virus (EBV), human immunodeficiency virus (HIV), and coronavirus (severe acute respiratory syndrome coronavirus 2 [SARS‐CoV‐2]) represent a great burden to human health worldwide. FDA‐approved anti‐parasite drug ivermectin is also an antibacterial, antiviral, and anticancer agent, which offers more potentiality to improve global public health, and it can effectively inhibit the replication of SARS‐CoV‐2 in vitro. This study sought to identify ivermectin‐related virus infection pathway alterations in human ovarian cancer cells. Stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics was used to analyze human ovarian cancer cells TOV‐21G treated with and without ivermectin (20 μmol/L) for 24 h, which identified 4447 ivermectin‐related proteins in ovarian cancer cells. Pathway network analysis revealed four statistically significant antiviral pathways, including HCMV, HPV, EBV, and HIV1 infection pathways. Interestingly, compared with the reported 284 SARS‐CoV‐2/COVID‐19‐related genes from GencLip3, we identified 52 SARS‐CoV‐2/COVID‐19‐related protein alterations when treated with and without ivermectin. Protein–protein network (PPI) was constructed based on the interactions between 284 SARS‐CoV‐2/COVID‐19‐related genes and between 52 SARS‐CoV‐2/COVID‐19‐related proteins regulated by ivermectin. Molecular complex detection analysis of PPI network identified three hub modules, including cytokines and growth factor family, MAP kinase and G‐protein family, and HLA class proteins. Gene Ontology analysis revealed 10 statistically significant cellular components, 13 molecular functions, and 11 biological processes. These findings demonstrate the broad‐spectrum antiviral property of ivermectin benefiting for COVID‐19 treatment in the context of predictive, preventive, and personalized medicine in virus‐related diseases.</jats:p>",
"alternative-id": [
"10.1002/jcp.30055"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2020-07-21"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2020-09-07"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2020-09-22"
}
],
"author": [
{
"affiliation": [
{
"name": "University Creative Research Initiatives Center Shandong First Medical University Jinan Shandong China"
},
{
"name": "Key Laboratory of Cancer Proteomics of Chinese Ministry of Health, Xiangya Hospital Central South University Changsha Hunan China"
},
{
"name": "State Local Joint Engineering Laboratory for Anticancer Drugs, Xiangya Hospital Central South University Changsha Hunan China"
}
],
"family": "Li",
"given": "Na",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Obstetrics and Gynecology, The Third Affiliated Hospital Sothern Medical University Tianhe Guangzhou Guangdong China"
}
],
"family": "Zhao",
"given": "Lingfeng",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4984-3549",
"affiliation": [
{
"name": "University Creative Research Initiatives Center Shandong First Medical University Jinan Shandong China"
},
{
"name": "Key Laboratory of Cancer Proteomics of Chinese Ministry of Health, Xiangya Hospital Central South University Changsha Hunan China"
},
{
"name": "State Local Joint Engineering Laboratory for Anticancer Drugs, Xiangya Hospital Central South University Changsha Hunan China"
},
{
"name": "Department of Oncology, Xiangya Hospital Central South University Changsha Hunan China"
},
{
"name": "National Clinical Research Center for Geriatric Disorders, Xiangya Hospital Central South University Changsha Hunan China"
}
],
"authenticated-orcid": false,
"family": "Zhan",
"given": "Xianquan",
"sequence": "additional"
}
],
"container-title": "Journal of Cellular Physiology",
"container-title-short": "Journal Cellular Physiology",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2020,
9,
22
]
],
"date-time": "2020-09-22T11:23:01Z",
"timestamp": 1600773781000
},
"deposited": {
"date-parts": [
[
2023,
8,
18
]
],
"date-time": "2023-08-18T22:13:29Z",
"timestamp": 1692396809000
},
"indexed": {
"date-parts": [
[
2024,
5,
13
]
],
"date-time": "2024-05-13T21:13:01Z",
"timestamp": 1715634781583
},
"is-referenced-by-count": 28,
"issue": "4",
"issued": {
"date-parts": [
[
2020,
9,
22
]
]
},
"journal-issue": {
"issue": "4",
"published-print": {
"date-parts": [
[
2021,
4
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://onlinelibrary.wiley.com/termsAndConditions#vor",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
9,
22
]
],
"date-time": "2020-09-22T00:00:00Z",
"timestamp": 1600732800000
}
}
],
"link": [
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1002/jcp.30055",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/full-xml/10.1002/jcp.30055",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1002/jcp.30055",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "311",
"original-title": [],
"page": "2959-2975",
"prefix": "10.1002",
"published": {
"date-parts": [
[
2020,
9,
22
]
]
},
"published-online": {
"date-parts": [
[
2020,
9,
22
]
]
},
"published-print": {
"date-parts": [
[
2021,
4
]
]
},
"publisher": "Wiley",
"reference": [
{
"DOI": "10.1007/s00436-019-06539-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_2_1"
},
{
"DOI": "10.1016/j.drudis.2019.07.011",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_3_1"
},
{
"DOI": "10.1016/j.mehy.2020.109882",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_4_1"
},
{
"DOI": "10.1111/j.1440-1711.2005.01412.x",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_5_1"
},
{
"DOI": "10.1016/j.ijantimicag.2019.05.003",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_6_1"
},
{
"DOI": "10.1016/j.bpobgyn.2020.02.009",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_7_1"
},
{
"DOI": "10.1186/1471-2105-4-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_8_1"
},
{
"DOI": "10.1093/bioinformatics/btp101",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_9_1"
},
{
"DOI": "10.1002/jmv.25688",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_10_1"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_11_1"
},
{
"article-title": "[Ivermectin]",
"author": "Boussinesq M.",
"first-page": "69",
"issue": "1",
"journal-title": "Medecine tropicale",
"key": "e_1_2_9_12_1",
"volume": "65",
"year": "2005"
},
{
"DOI": "10.1007/978-3-540-77349-8_23",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_13_1"
},
{
"DOI": "10.1046/j.1464-410X.2002.02962.x",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_14_1"
},
{
"DOI": "10.1128/AAC.15.3.361",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_15_1"
},
{
"article-title": "Primary human cytomegalovirus (HCMV) infection in pregnancy",
"author": "Buxmann H.",
"first-page": "45",
"issue": "4",
"journal-title": "Deutsches Ärzteblatt International",
"key": "e_1_2_9_16_1",
"volume": "114",
"year": "2017"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_17_1"
},
{
"DOI": "10.1021/jm00184a014",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_18_1"
},
{
"DOI": "10.4269/ajtmh.20-0271",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_19_1"
},
{
"DOI": "10.1038/ja.2017.11",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_20_1"
},
{
"DOI": "10.1172/jci.insight.99431",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_21_1"
},
{
"DOI": "10.1002/jcb.27490",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_22_1"
},
{
"DOI": "10.1016/S0151-9638(04)93668-X",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_23_1"
},
{
"DOI": "10.1186/s12917-019-2026-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_24_1"
},
{
"DOI": "10.1158/0008-5472.CAN-15-2887",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_25_1"
},
{
"DOI": "10.2147/DDDT.S172538",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_26_1"
},
{
"DOI": "10.1177/009127002237994",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_27_1"
},
{
"DOI": "10.1002/path.2067",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_28_1"
},
{
"DOI": "10.1002/JLB.2AB0418-158RR",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_29_1"
},
{
"DOI": "10.1016/j.tim.2017.07.007",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_30_1"
},
{
"author": "Huynh K.",
"key": "e_1_2_9_31_1",
"volume-title": "HIV prevention",
"year": "2020"
},
{
"DOI": "10.1186/s13046-019-1251-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_32_1"
},
{
"article-title": "The multitargeted drug ivermectin: From an antiparasitic agent to a repositioned cancer drug",
"author": "Juarez M.",
"first-page": "317",
"issue": "2",
"journal-title": "American Journal of Cancer Research",
"key": "e_1_2_9_33_1",
"volume": "8",
"year": "2018"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105924",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_34_1"
},
{
"DOI": "10.1016/j.pt.2017.02.004",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_35_1"
},
{
"DOI": "10.1016/j.pestbp.2019.06.009",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_36_1"
},
{
"DOI": "10.1038/s41564-020-0688-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_37_1"
},
{
"DOI": "10.1111/bjd.18369",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_38_1"
},
{
"DOI": "10.1128/AAC.00483-20",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_39_1"
},
{
"DOI": "10.1007/s13167-020-00209-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_40_1"
},
{
"DOI": "10.1016/j.antiviral.2018.09.010",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_41_1"
},
{
"DOI": "10.1016/j.pathol.2019.09.006",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_42_1"
},
{
"DOI": "10.1007/s11904-019-00452-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_43_1"
},
{
"DOI": "10.1016/S2214-109X(19)30453-X",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_44_1"
},
{
"DOI": "10.1177/000992280304200702",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_45_1"
},
{
"DOI": "10.1016/j.jaad.2020.04.017",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_46_1"
},
{
"DOI": "10.1021/ac026117i",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_47_1"
},
{
"DOI": "10.1038/s41577-020-0288-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_48_1"
},
{
"DOI": "10.1016/j.humpath.2018.05.020",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_49_1"
},
{
"DOI": "10.1111/jdv.14437",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_50_1"
},
{
"DOI": "10.1016/j.coviro.2013.12.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_51_1"
},
{
"DOI": "10.1093/nar/gku1003",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_52_1"
},
{
"DOI": "10.1007/s12015-020-09976-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_53_1"
},
{
"DOI": "10.1136/vr.123.9.226",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_54_1"
},
{
"DOI": "10.1016/j.antiviral.2015.12.012",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_55_1"
},
{
"DOI": "10.1042/BJ20120150",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_56_1"
},
{
"DOI": "10.1093/bioinformatics/btz807",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_57_1"
},
{
"DOI": "10.1016/j.dci.2020.103744",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_58_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104760",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_59_1"
},
{
"DOI": "10.1089/omi.2011.0118",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_60_1"
},
{
"DOI": "10.1016/S0378-3782(14)70010-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_61_1"
},
{
"DOI": "10.1128/JVI.01952-13",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_62_1"
},
{
"DOI": "10.1016/j.intimp.2008.12.016",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_63_1"
}
],
"reference-count": 62,
"references-count": 62,
"relation": {},
"resource": {
"primary": {
"URL": "https://onlinelibrary.wiley.com/doi/10.1002/jcp.30055"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Quantitative proteomics reveals a broad‐spectrum antiviral property of ivermectin, benefiting for COVID‐19 treatment",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy",
"volume": "236"
}