Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach
et al., Biotecnia, doi:10.18633/biotecnia.v27.2485, Mar 2025
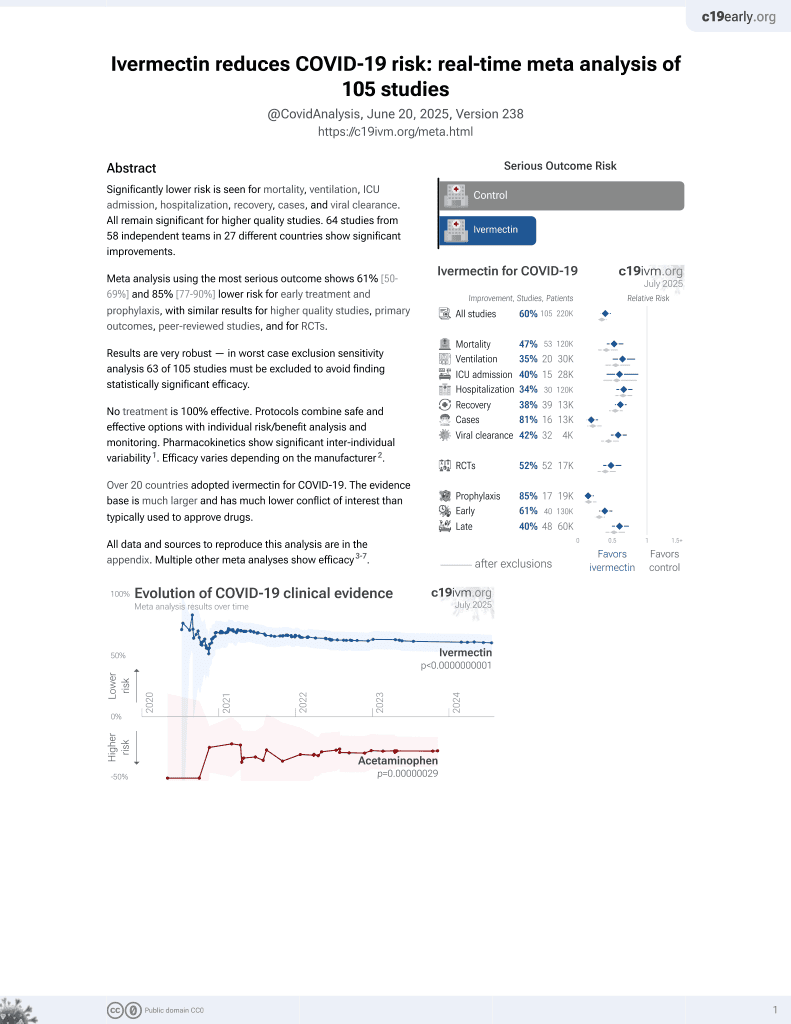
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico study showing that ivermectin, the SARS-CoV-2 nucleocapsid (N) protein, and the ORF6 protein share binding sites on human importin α isoforms. Authors used molecular docking to analyze binding affinities between these molecules and various importin α isoforms, finding that all three bind to the ARM2-ARM4 domains (major binding site) with favorable binding energies. Ivermectin may compete with viral proteins for binding to importin α, potentially explaining antiviral activity against SARS-CoV-2. Specifically, ivermectin showed binding affinities to importin α isoforms with free binding energies ranging from -7.71 to -8.63 kcal/mol, comparable to those of viral proteins. This suggests ivermectin could interfere with viral protein transport into the nucleus and inhibit the virus's ability to suppress host interferon-mediated antiviral responses.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Gayozo et al., 13 Mar 2025, USA, peer-reviewed, 3 authors.
Contact: elviologo@gmail.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach
Biotecnia, doi:10.18633/biotecnia.v27.2485
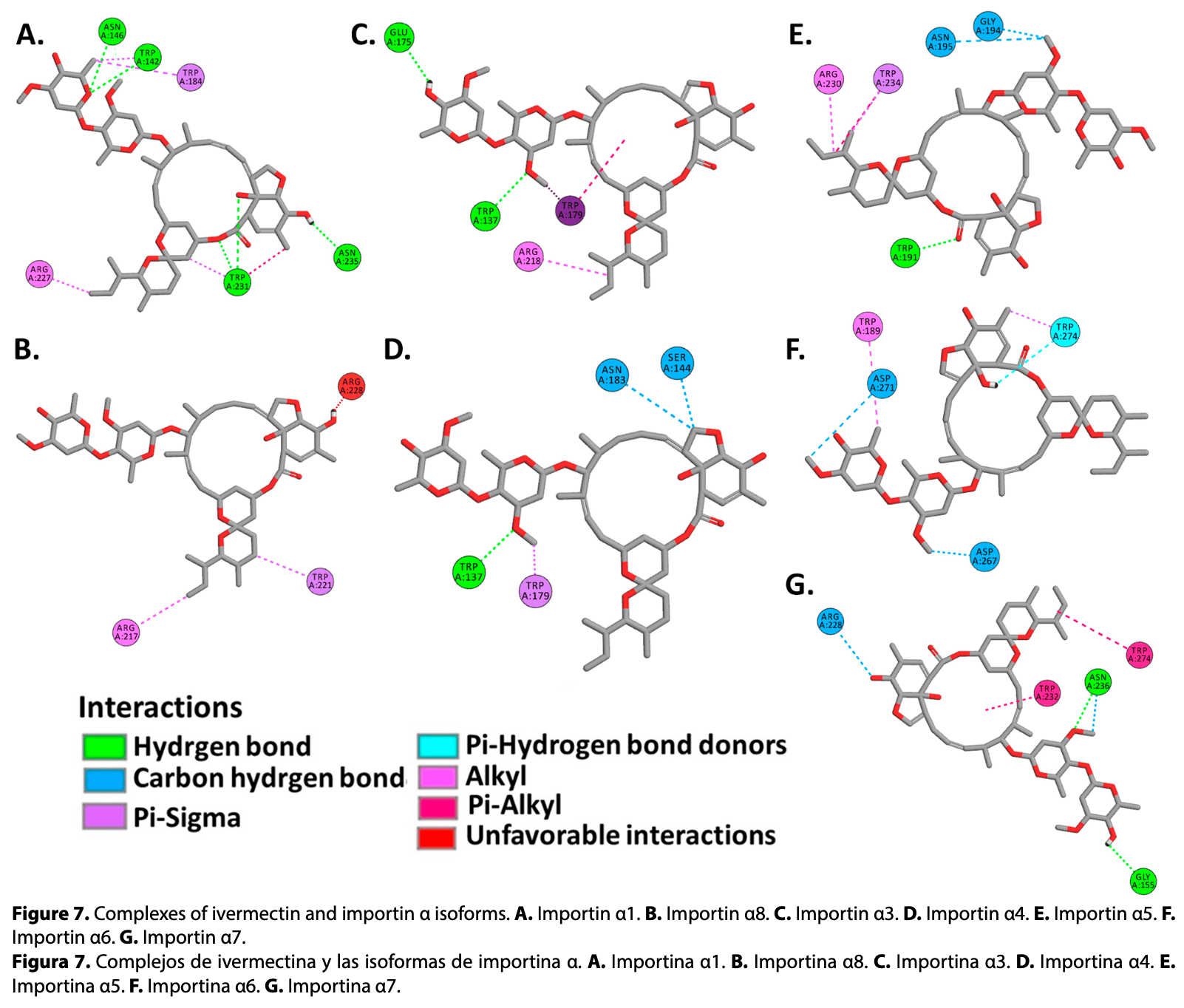
Ivermectin has been shown in vitro that reduces SARS-CoV-2 replication in infected cells through interactions with importins α, however, the exact mechanism of action is still unknown. The objective of this study was to analyze binding affinities of ivermectin, SARS-CoV-2 nucleocapsid (N) and ORF6 proteins, to isoforms of human importins α using molecular docking methods. Crystallized structures of importins α from Protein Data Bank (PDB) and AlphaFold Protein Structure Database were used, viral proteins were modeled using AlphaFold 2. Molecular docking simulations were performed between human importin α isoforms, ivermectin, N and ORF6 proteins, employing Broyden-Fletcher-Goldfarb-Shanno, FTDock and pyDockRST algorithms. Obtained data evidenced that viral proteins of SARS-CoV-2 and ivermectin showed favorable binding affinities to ARM2-ARM4 domains (major binding site), sharing binding affinities to the same active residues. These results suggest that ivermectin shares the same active site on the α-importins as the SARS-CoV-2 N and ORF6 proteins, demonstrating a potential molecular target for research in the development of new antiviral drugs against COVID-19.
Resulting complexes between importins α isoforms and N protein (NLS pat4 and pat7) showed free binding energy values (∆G) ranging -10.0 to -6.3 kcal.mol -1 , where the NLS pat7 sequence demonstrated the most favorable energy value, specifically to importin α3 and importin α5 (Figure 3 .B, Supplementary Table S1 ). Active residues in complexes formed by N protein and isoforms of α1 subfamily were Phe138, Trp142, Asn146, Ser149, His177, Glu180, Trp184, Asn188, Asn228 in importin α1, and Arg95, Gln100, Glu107, Trp136, Ser143, Glu180, Asn182, Trp231, Glu256, Asp260, Glu266, Asp270, Trp273 in importin α8 , residues that interact with NLS pat4 sequences through hydrogen bonds, unconventional interactions between a polarized carbon atom and hydrogen atom, interactions between Pi orbitals and donors groups of hydrogen bonds. The formation of electrostatic attractions and hydrophobic interactions were also identified (Figure 3 .A,B, Supplementary Figure S2 , Supplementary Figure S3 , Supplementary Table S1 ). Residues Glu107, Trp142, Asn146, Ala176, Glu180, Trp184, Ala222, Tyr225, Trp231, Glu266, Asp270, Trp273 of importin α1, and residues Ser99, Glu101, Pro104, Trp136, Ser143, Arg217, Asp260, Trp263, Glu174, Trp178, Asn182, Trp221, Asn225 of importin α8, interact by hydrogen bonds, carbon hydrogen bonds, Pi interactions with hydrogen bonds donors, hydrophobic and electrostatic interactions with the NLS pat7 sequences (Figure 4 .A,B, Supplementary Figure S2 , Supplementary..
References
Addetia, Lieberman, Phung, Hsiang, Xie et al., SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98, mBio, doi:10.1128/mBio.00065-21
Andersen, Rambaut, Lipkin, Holmes, Garry, The proximal origin of SARS-CoV-2, Nature Medicine, doi:10.1038/s41591-020-0820-9
Azam, Taban, Eid, Iqbal, Alam et al., An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.184102
Baumhardt, Chook, Structures of Importins and Exportins, doi:10.1007/978-3-319-77309-4_6
Bello, Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857
Berman, Westbrook, Feng, Gilliland, Bhat et al., The protein data bank, Nucleic Acids Res, doi:10.1093/nar/28.1.235
Bian, Wilson, Common importin alpha specificity for papillomavirus E2 proteins, Virus Research, doi:10.1016/j.virusres.2010.02.011
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787
Ceraolo, Giorgi, Genomic variance of the 2019-nCoV coronavirus, Journal of Medical Virology, doi:10.1002/jmv.25700
Chang, Hou, Chang, Hsiao, Huang, The SARS coronavirus nucleocapsid protein -Forms and functions, Antiviral Research, doi:10.1016/j.antiviral.2013.12.009
Chelliah, Blundell, Fernández-Recio, Efficient restraints for protein-protein docking by comparison of observed amino acid substitution patterns with those predicted from local Environment, Journal of Molecular Biology, doi:10.1016/j.jmb.2006.01.001
Chen, Boon, Wang, Chan, Chan, Genomic and evolutionary comparison between SARS-CoV-2 and other human coronaviruses, Journal of Virological Methods, doi:10.1016/j.jviromet.2020.114032
Cheng, Blundell, Fernandez-Recio, pyDock: Electrostatics and desolvation for effective scoring of rigid-body protein-protein docking, Proteins: Structure, Function, and Bioinformatics, doi:10.1002/prot.21419
Chook, Blobel, Karyopherins and nuclear import, Current Opinion in Structural Biology, doi:10.1016/S0959-440X(01)00264-0
Choudhury, Das, Patra, Bhattacharya, Ghosh et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Virology, doi:10.2217/fvl-2020-0342
Crump, Ivermectin: enigmatic multifaceted 'wonder' drug continues to surprise and exceed expectations, The Journal of Antibiotics, doi:10.1038/ja.2017
Dallakyan, Olson, Small-molecule library screening by docking with PyRx, doi:10.1007/978-1-4939-2269-7_19
David, Islam, Tankhilevich, Sternberg, The AlphaFold database of protein structures: A Biologist's Guide, Journal of Molecular Biology, doi:10.1016/j.jmb.2021.167336
Fagerlund, Melén, Kinnunen, Julkunen, Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and Importin α5, Journal of Biological Chemistry, doi:10.1074/jbc.M202943200
Fang, Jang, Watson, Wellappili, Tyler, Distinctive nuclear localization signals in the oomycete Phytophthora sojae, Front. Microbiol, doi:10.3389/fmicb.2017.00010
Fontes, Teh, Kobe, Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11, Journal of Molecular Biology, doi:10.1006/jmbi.2000.3642
Frieman, Yount, Heise, Kopecky-Bromberg, Palese et al., Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane, Journal of Virology, doi:10.1128/JVI.01012-07
Fry, Saladi, Cunha, Clemons, Sequence-based features that are determinant for tailanchored membrane protein sorting in eukaryotes, Traffic, doi:10.1111/tra.12809
Gabb, Jackson, Sternberg, Modelling protein docking using shape complementarity, electrostatics and biochemical information, J. Thornton. Journal of Molecular Biology, doi:10.1006/jmbi.1997.1203
Gao, Gao, Liu, Nie, Sun et al., Identification and functional analysis of the SARS-COV-2 nucleocapsid protein, BMC Microbiology, doi:10.1186/s12866-021-02107-3
Gayozo, Rojas, Interacción in silico de las moléculas Agathisflavona, Amentoflavona y Punicalina con la Importina α1 humana, Revista Colombiana de Biotecnología, doi:10.15446/rev.colomb.biote.v23n2.94466
González-Paz, Hurtado-León, Lossada, Fernández-Materán, Vera-Villalobos et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677
Gordon, Jang, Bouhaddou, Xu, Obernier et al., None
Grosdidier, Pons, Solernou, Fernández-Recio, Prediction and scoring of docking poses with pyDock, Proteins: Structure, Function, and Bioinformatics, doi:10.1002/prot.21796
Gupta, Biswal, Panda, Ray, Rana, Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1839564
Hanwell, Curtis, Lonie, Vandermeersch, Zurek et al., Avogadro: an advanced semantic chemical editor, visualization, and analysis platform, J. Cheminform, doi:10.1186/1758-2946-4-17
He, Dobie, Ballantine, Leeson, Li et al., Analysis of multimerization of the SARS coronavirus nucleocapsid protein, Biochemical and Biophysical Research Communications, doi:10.1016/j.bbrc.2004.02.074
Heo, Lee, Seok, GalaxyRefineComplex: Refinement of protein-protein complex model structures driven by interface repacking, Sci Rep, doi:10.1038/srep32153
Horton, Park, Obayashi, Fujita, Harada et al., WoLF PSORT: protein localization predictor, Nucleic Acids Res, doi:10.1093/nar/gkm259
Hussain, Gallagher, SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus, Virus Research, doi:10.1016/j.virusres.2010.08.017
Ibrahim, Abdelmalek, Elshahat, Elfiky, COVID-19 spike-host cell receptor GRP78 binding site prediction, Journal of Infection, doi:10.1016/j.jinf.2020.02.026
Iqbal, Romero-Castillo, Bilal, Parra-Saldivar, The emergence of novel-coronavirus and its replication cycle -An overview, Journal of Pure and Applied Microbiology, doi:10.22207/JPAM.14.1.03
Jiménez-García, Pons, Fernández-Recio, pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring, Bioinformatics, doi:10.1093/bioinformatics/btt262
Jumper, Evans, Pritzel, Green, Figurnov et al., Highly accurate protein structure prediction with AlphaFold, Nature, doi:10.1038/s41586-021-03819-2
Kannan, Shaik Syed Ali, Sheeza, Hemalatha, COVID-19 (Novel Coronavirus 2019) -recent trends, Eur Rev Med Pharmacol Sci, doi:10.26355/eurrev_202002_20378
Kato, Ikliptikawati, Kobayashi, Kondo, Lim et al., Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex, Biochemical and Biophysical Research Communications, doi:10.1016/j.bbrc.2020.11.115
Kim, Thiessen, Bolton, Chen, Fu et al., PubChem substance and compound databases, Nucleic Acids Res, doi:10.1093/nar/gkv951
King, Tessier, Dodge, Weinberg, Mymryk, Inhibition of human adenovirus replication by the importin α/β1 nuclear import inhibitor ivermectin, Journal of Virology, doi:10.1128/JVI.00710-20
Kopecky-Bromberg, Martínez-Sobrido, Frieman, Baric, Palese, Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists, Journal of Virology, doi:10.1128/JVI.01782-06
Kumar, Stecher, Li, Knyaz, Tamura, MEGA X: Molecular evolutionary genetics analysis across computing platforms, Mol Biol Evol, doi:10.1093/molbev/msy096
Kyte, Doolittle, A simple method for displaying the hydropathic character of a protein, Journal of Molecular Biology, doi:10.1016/0022-2836(82)90515-0
Laskowski, Macarthur, Moss, Thornton, PROCHECK: a program to check the stereochemical quality of protein structures, J. Appl. Cryst, doi:10.1107/S0021889892009944
Li, Cowley, Uludag, Gur, Mcwilliam et al., The EMBL-EBI bioinformatics web and programmatic tools framework, Nucleic Acids Res, doi:10.1093/nar/gkv279
Li, Liao, Wang, Tan, Luo et al., The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway, Virus Research, doi:10.1016/j.virusres.2020.198074
Liu, Fung, Chong, Shukla, Hilgenfeld, Accessory proteins of SARS-CoV and other coronaviruses, Antiviral Research, doi:10.1016/j.antiviral.2014.06.013
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, The Lancet, doi:10.1016/S0140-6736(20)30251-8
Mastrangelo, Pezzullo, De Burghgraeve, Kaptein, Pastorino et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, J. Antimicrob Chemother, doi:10.1093/jac/dks147
Mirdita, Schütze, Moriwaki, Heo, Ovchinnikov et al., ColabFold: making protein folding accessible to all, Nat Methods, doi:10.1038/s41592-022-01488-1
Miyamoto, Itoh, Suzuki, Tanaka, Sakai et al., SARS-CoV-2 ORF6 disrupts nucleocytoplasmic trafficking to advance viral replication, Commun Biol, doi:10.1038/s42003-022-03427-4
Narayanan, Huang, Makino, SARS coronavirus accessory proteins, Virus Research, SARS-CoV Pathogenesis and Replication, doi:10.1016/j.virusres.2007.10.009
Pallara, Jiménez-García, Romero, Moal, Fernández-Recio, pyDock scoring for the new modeling challenges in docking: Protein-peptide, homomultimers, and domain-domain interactions, Proteins: Structure, Function, and Bioinformatics, doi:10.1002/prot.25184
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera-A visualization system for exploratory research and analysis, Journal of Computational Chemistry, doi:10.1002/jcc.20084
Pumroy, Cingolani, Diversification of importin-α isoforms in cellular trafficking and disease states, Biochemical Journal, doi:10.1042/BJ20141186
Pumroy, Ke, Hart, Zachariae, Cingolani, Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7, Structure, doi:10.1016/j.str.2014.11.015
Qinfen, Jinming, Xiaojun, Huanying, Jicheng et al., The life cycle of SARS coronavirus in Vero E6 cells, Journal of Medical Virology, doi:10.1002/jmv.20095
Rizzo, Ivermectin, antiviral properties and COVID-19: a possible new mechanism of action, Naunyn-Schmiedeberg's Arch Pharmacol, doi:10.1007/s00210-020-01902-5
Robert, Gouet, Deciphering key features in protein structures with the new ENDscript server, Nucleic Acids Res, doi:10.1093/nar/gku316
Rowland, Chauhan, Fang, Pekosz, Kerrigan et al., Intracellular localization of the severe acute respiratory syndrome Ccoronavirus nucleocapsid protein: Absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells, Journal of Virology, doi:10.1128/JVI.79.17.11507-11512.2005
Saha, Raihan, The binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Struct Chem, doi:10.1007/s11224-021-01776-0
Sekimoto, Imamoto, Nakajima, Hirano, Yoneda, Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1, The EMBO Journal, doi:10.1093/emboj/16.23.7067
Shereen, Khan, Kazmi, Bashir, Siddique, COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses, Journal of Advanced Research, doi:10.1016/j.jare.2020.03.005
Sievers, Higgins, Clustal omega, accurate alignment of very large numbers of sequences, doi:10.1007/978-1-62703-646-7_6
Surjit, Liu, Jameel, Chow, Lal, The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors, Biochem J, doi:10.1042/BJ20040984
Tarendeau, Boudet, Guilligay, Mas, Bougault et al., Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit, Nat Struct Mol Biol, doi:10.1038/nsmb1212
Timani, Liao, Ye, Linbai, Zeng et al., Nuclear/ nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus, Virus Research, doi:10.1016/j.virusres.2005.05.007
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, Journal of Computational Chemistry, doi:10.1002/jcc.21334
Vangone, Schaarschmidt, Koukos, Geng, Citro et al., Large-scale prediction of binding affinity in protein-small ligand complexes: the PRODIGY-LIG web server, Bioinformatics, doi:10.1093/bioinformatics/bty816
Varadi, Anyango, Deshpande, Nair, Natassia et al., AlphaFold Protein Structure Database: massively expanding the structural coverage of proteinsequence space with high-accuracy models, Nucleic Acids Research, doi:10.1093/nar/gkab1061
Wagstaff, Sivakumaran, Heaton, Harrich, Jans, Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150
Williams, Headd, Moriarty, Prisant, Videau et al., MolProbity: More and better reference data for improved all-atom structure validation, Protein Science, doi:10.1002/pro.3330
Wu, Zhao, Yu, Chen, Wang et al., A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Wulan, Heydet, Walker, Gahan, Ghildyal, Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses, Front. Microbiol, doi:10.3389/fmicb.2015.00553
Wurm, Chen, Hodgson, Britton, Brooks et al., Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division, Journal of Virology, doi:10.1128/JVI.75.19.9345-9356.2001
Xu, Edwards, Borek, Feagins, Mittal et al., Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1, Cell Host & Microbe, doi:10.1016/j.chom.2014.07.008
Yang, Atkinson, Wang, Lee, Bogoyevitch et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760
Ye, Wong, Li, Xie, A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNKdependent apoptosis, Biochimica et Biophysica Acta (BBA) -General Subjects, doi:10.1016/j.bbagen.2008.07.009
Zaidi, Dehgani-Mobaraki, The mechanisms of action of ivermectin against SARS-CoV-2-an extensive review, J Antibiot, doi:10.1038/s41429-021-00491-6
Zhao, Falcón, Zhou, Netland, Enjuanes et al., Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication, Journal of Virology, doi:10.1128/JVI.02371-08
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Ōmura, Crump, Ivermectin: panacea for resourcepoor communities?, Trends in Parasitology, doi:10.1016/j.pt.2014.07.005
DOI record:
{
"DOI": "10.18633/biotecnia.v27.2485",
"ISSN": [
"1665-1456",
"1665-1456"
],
"URL": "http://dx.doi.org/10.18633/biotecnia.v27.2485",
"abstract": "<jats:p>Ivermectin has been shown in vitro that reduces SARS-CoV-2 replication in infected cells through interactions with importins α, however, the exact mechanism of action is still unknown. The objective of this study was to analyze binding affinities of ivermectin, SARS-CoV-2 nucleocapsid (N) and ORF6 proteins, to isoforms of human importins α using molecular docking methods. Crystallized structures of importins α from Protein Data Bank (PDB) and AlphaFold Protein Structure Database were used, viral proteins were modeled using AlphaFold 2. Molecular docking simulations were performed between human importin α isoforms, ivermectin, N and ORF6 proteins, employing Broyden-Fletcher-Goldfarb-Shanno, FTDock and pyDockRST algorithms. Data obtained evidenced that viral proteins of SARS-CoV-2 and ivermectin showed favorable binding affinities to ARM2-ARM4 domains (major binding site), sharing binding affinities to the same active residues. These results suggest that ivermectin shares the same active site on the α-importins as the SARS-CoV-2 N and ORF6 proteins, demonstrating a potential molecular target for research in the development of new antiviral drugs against COVID-19.</jats:p>",
"author": [
{
"ORCID": "https://orcid.org/0000-0001-9309-7056",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gayozo",
"given": "Elvio",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-8341-7746",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rojas",
"given": "Laura",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4120-4141",
"affiliation": [],
"authenticated-orcid": false,
"family": "Barrios",
"given": "Julio",
"sequence": "additional"
}
],
"container-title": "Biotecnia",
"container-title-short": "BIOTECNIA",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
3,
18
]
],
"date-time": "2025-03-18T20:53:10Z",
"timestamp": 1742331190000
},
"deposited": {
"date-parts": [
[
2025,
3,
18
]
],
"date-time": "2025-03-18T20:53:30Z",
"timestamp": 1742331210000
},
"indexed": {
"date-parts": [
[
2025,
3,
19
]
],
"date-time": "2025-03-19T04:33:22Z",
"timestamp": 1742358802766,
"version": "3.40.1"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
3,
13
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-sa/4.0",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T00:00:00Z",
"timestamp": 1741824000000
}
}
],
"link": [
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1391",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1392",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1391",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "7961",
"original-title": [],
"page": "e2485",
"prefix": "10.18633",
"published": {
"date-parts": [
[
2025,
3,
13
]
]
},
"published-online": {
"date-parts": [
[
2025,
3,
13
]
]
},
"publisher": "Universidad de Sonora",
"reference": [
{
"DOI": "10.1128/mBio.00065-21",
"doi-asserted-by": "crossref",
"key": "75797",
"unstructured": "Addetia, A., Lieberman, N. A. P., Phung, Q., Hsiang, T.Y., Xie, H., Roychoudhury, P., Shrestha, L., Loprieno, M. A, Huang, M. L., Gale, M. J., Jerome, K. R. and Greninger, A. L. 2021. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio, 12(2), e00065-21. https://doi.org/10.1128/mBio.00065-21"
},
{
"DOI": "10.1038/s41591-020-0820-9",
"doi-asserted-by": "crossref",
"key": "75798",
"unstructured": "Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C. and Garry, R. F. 2020. The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450-452. https://doi.org/10.1038/s41591-020-0820-9"
},
{
"DOI": "10.1080/07391102.2020.1841028",
"doi-asserted-by": "crossref",
"key": "75799",
"unstructured": "Azam, F., Taban, I. M., Eid, E. E. M., Iqbal, M., Alam, O., Khan, S., Mahmood, D., Anwar, M. J., Khaililullah, H. and Khan, M. U. 2020. An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α. Journal of Biomolecular Structure and Dynamics, 40(6), 2851–2864. https://doi.org/10.1080/07391102.2020.1841028"
},
{
"DOI": "10.1007/978-3-319-77309-4_6",
"doi-asserted-by": "crossref",
"key": "75800",
"unstructured": "Baumhardt, J. and Chook, Y. M. 2018. Structures of Importins and Exportins. En W. Yang (Ed.), Nuclear-Cytoplasmic Transport (pp. 113-149). Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-77309-4_6"
},
{
"DOI": "10.1080/07391102.2021.1911857",
"doi-asserted-by": "crossref",
"key": "75801",
"unstructured": "Bello, M. 2022. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. Journal of Biomolecular Structure and Dynamics, 40(18), 8375-8383. https://doi.org/10.1080/07391102.2021.1911857"
},
{
"DOI": "10.1093/nar/28.1.235",
"doi-asserted-by": "crossref",
"key": "75802",
"unstructured": "Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. and Bourne, P. E. 2000. The Protein Data Bank. Nucleic Acids Research, 28(1), 235-242. https://doi.org/10.1093/nar/28.1.235"
},
{
"DOI": "10.1016/j.virusres.2010.02.011",
"doi-asserted-by": "crossref",
"key": "75803",
"unstructured": "Bian, X.L. and Wilson, V. G. 2010. Common importin alpha specificity for papillomavirus E2 proteins. Virus Research, 150(1), 135-137. https://doi.org/10.1016/j.virusres.2010.02.011"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "crossref",
"key": "75804",
"unstructured": "Caly, L., Druce, J. D., Catton, M. G., Jans, D. A. and Wagstaff, K. M. 2020. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. https://doi.org/10.1016/j.antiviral.2020.104787"
},
{
"DOI": "10.1002/jmv.25700",
"doi-asserted-by": "crossref",
"key": "75805",
"unstructured": "Ceraolo, C. and Giorgi, F. M. 2020. Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92(5), 522-528. https://doi.org/10.1002/jmv.25700"
},
{
"DOI": "10.1016/j.antiviral.2013.12.009",
"doi-asserted-by": "crossref",
"key": "75806",
"unstructured": "Chang, C., Hou, M.H., Chang, C.F., Hsiao, C.D. and Huang, T. 2014. The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research, 103, 39-50. https://doi.org/10.1016/j.antiviral.2013.12.009"
},
{
"DOI": "10.1016/j.jmb.2006.01.001",
"doi-asserted-by": "crossref",
"key": "75807",
"unstructured": "Chelliah, V., Blundell, T. L. and Fernández-Recio, J. 2006. Efficient Restraints for Protein–Protein Docking by Comparison of Observed Amino Acid Substitution Patterns with those Predicted from Local Environment. Journal of Molecular Biology, 357(5), 1669-1682. https://doi.org/10.1016/j.jmb.2006.01.001"
},
{
"DOI": "10.1002/prot.21419",
"doi-asserted-by": "crossref",
"key": "75808",
"unstructured": "Cheng, T. M.K., Blundell, T. L. and Fernandez-Recio, J. 2007. pyDock: Electrostatics and desolvation for effective scoring of rigid-body protein–protein docking. Proteins: Structure, Function, and Bioinformatics, 68(2), 503-515. https://doi.org/10.1002/prot.21419"
},
{
"DOI": "10.1016/S0959-440X(01)00264-0",
"doi-asserted-by": "crossref",
"key": "75809",
"unstructured": "Chook, Y. and Blobel, G. 2001. Karyopherins and nuclear import. Current Opinion in Structural Biology, 11(6), 703-715. https://doi.org/10.1016/S0959-440X(01)00264-0"
},
{
"DOI": "10.2217/fvl-2020-0342",
"doi-asserted-by": "crossref",
"key": "75810",
"unstructured": "Choudhury, A., Das, N. C., Patra, R., Bhattacharya, M., Ghosh, P., Patra, B. C. and Mukherjee, S. 2021. Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach. Future Virology, 16(4), 277-291. https://doi.org/10.2217/fvl-2020-0342"
},
{
"DOI": "10.1038/ja.2017.11",
"doi-asserted-by": "crossref",
"key": "75811",
"unstructured": "Crump, A. 2017. Ivermectin: Enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. The Journal of Antibiotics, 70(5), 495-505. https://doi.org/10.1038/ja.2017.11"
},
{
"DOI": "10.1007/978-1-4939-2269-7_19",
"doi-asserted-by": "crossref",
"key": "75812",
"unstructured": "Dallakyan, S. and Olson, A. J. 2015. Small-Molecule Library Screening by Docking with PyRx. En J. E. Hempel, C. H. Williams, and C. C. Hong (Eds.), Chemical Biology: Methods and Protocols (pp. 243-250). New York, NY: Springer. https://doi.org/10.1007/978-1-4939-2269-7_19"
},
{
"DOI": "10.1016/j.jmb.2021.167336",
"doi-asserted-by": "crossref",
"key": "75813",
"unstructured": "David, A., Islam, S., Tankhilevich, E. and Sternberg, M. J. E. 2022. The AlphaFold Database of Protein Structures: A Biologist’s Guide. Journal of Molecular Biology, 434(2), 167336. https://doi.org/10.1016/j.jmb.2021.167336"
},
{
"DOI": "10.1074/jbc.M202943200",
"doi-asserted-by": "crossref",
"key": "75814",
"unstructured": "Fagerlund, R., Melén, K., Kinnunen, L. and Julkunen, I. 2002. Arginine/Lysine-rich Nuclear Localization Signals Mediate Interactions between Dimeric STATs and Importin α5 *. Journal of Biological Chemistry, 277(33), 30072-30078. https://doi.org/10.1074/jbc.M202943200"
},
{
"DOI": "10.1006/jmbi.2000.3642",
"doi-asserted-by": "crossref",
"key": "75815",
"unstructured": "Fontes, M. R. M., Teh, T. and Kobe, B. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11Edited by K. Nagai. Journal of Molecular Biology, 297(5), 1183-1194. https://doi.org/10.1006/jmbi.2000.3642"
},
{
"DOI": "10.1128/JVI.01012-07",
"doi-asserted-by": "crossref",
"key": "75816",
"unstructured": "Frieman, M., Yount, B., Heise, M., Kopecky-Bromberg, S. A., Palese, P. and Baric, R. S. 2007. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. Journal of Virology, 81(18), 9812-9824. https://doi.org/10.1128/JVI.01012-07"
},
{
"DOI": "10.1006/jmbi.1997.1203",
"doi-asserted-by": "crossref",
"key": "75817",
"unstructured": "Gabb, H. A., Jackson, R. M. and Sternberg, M. J. E. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information11Edited by J. Thornton. Journal of Molecular Biology, 272(1), 106-120. https://doi.org/10.1006/jmbi.1997.1203"
},
{
"DOI": "10.1186/s12866-021-02107-3",
"doi-asserted-by": "crossref",
"key": "75818",
"unstructured": "Gao, T., Gao, Y., Liu, X., Nie, Z., Sun, H., Lin, K., Peng, H. and Wang, S. 2021. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC Microbiology, 21(1), 58. https://doi.org/10.1186/s12866-021-02107-3"
},
{
"DOI": "10.56152/ffs.v12i2.2219",
"doi-asserted-by": "crossref",
"key": "75819",
"unstructured": "Gayozo, E., Rojas, L. and López, M. 2020. Análisis computacional de la interacción in silico de la Agatisflavona con la región de trimerización de la proteína espícula (S) del SARS-CoV-2. Steviana, 12(2), 70-80. https://doi.org/10.56152/StevianaFacenV12N2A4_2020"
},
{
"DOI": "10.15446/rev.colomb.biote.v23n2.94466",
"doi-asserted-by": "crossref",
"key": "75820",
"unstructured": "Gayozo, E. and Rojas, L. 2021. Interacción in silico de las moléculas Agathisflavona, Amentoflavona y Punicalina con la Importina α1 humana. Revista Colombiana de Biotecnología, 23(2), 15-24. https://doi.org/10.15446/rev.colomb.biote.v23n2.94466"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "crossref",
"key": "75821",
"unstructured": "Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O’Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., Kim, M., Haas, P., Polacco, B. J., Braberg, H., Fabius, J. M., Eckhardt, M., Soucheray, M., Bennett, M. J., Cakir, M., McGregor, M. J., Li, Q., Meyer, B., Roesch, F., Vallet, T., Mac Kain, A., Miorin, L., Moreno, E., Chi Naing, Z. Z., Zhou, Y., Peng, S., Shi, Y., Zhang, Z., Shen, W., Kirby, I. T., Melnyk, J. E., Chorba, J. S., Lou, K., Dai, S. A., Barrio-Hernandez, I., Memon, D., Hernandez-Armenta, C., Lyu, J., Mathy, C. P., Perica, T., Bharath Pilla, K., Ganesan, S. J., Saltzberg, D. J., Rakesh, R., Liu, X., Rosenthal, S. B., Calviello, L., Venkataramanan, S., Liboy-Lugo, J., Lin, Y., Huang, X. P., Liu, Y., Wankowicz, S. A., Bohn, M., Safari, M., Ugur, F. S., Koh, C., Savar, N. S., Dinh Tran, Q., Shengjuler,D., Fletcher, S. J., O’Neal, M. C., Cai, Y., Chang, J. C., Broadhurst, D. J., Klippsten, S., Sharp, P. P., Wenzell, N. A., Kuzuoglu-Ozturk, D., Wang, H., Trenker, R., Young, J. M., Cavero, D. A., Hiatt, J., Roth, T. L., Rathore, U., Subramanian, A., Noack, J., Hubert, M., Stroud, R. M., Frankel, A. D., Rosenberg, O. S., Verba, K. A., Agard, D. A., Ott, M., Emerman, M., Jura, N., von Zastrow, M., Verdin, E., Ashworth, A., Schwartz, O., d’Enfert, C., Mukherjee, S., Jacobson, M., Malik, H. M., Fujimori, D. G., Ideker, T., Craik, C. S., Floor, S. N., Fraser, J. S., Gross, J. D., Sali, A., Roth, B. L., Ruggero, D., Taunton, J., Kortemme, T., Beltrao, P., Vignuzzi, M., García-Sastre, A., Shokat, K. M., Shoichet, B. K. and Krogan, N. J. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459-468. https://doi.org/10.1038/s41586-020-2286-9"
},
{
"DOI": "10.1002/prot.21796",
"doi-asserted-by": "crossref",
"key": "75822",
"unstructured": "Grosdidier, S., Pons, C., Solernou, A. and Fernández-Recio, J. 2007. Prediction and scoring of docking poses with pyDock. Proteins: Structure, Function, and Bioinformatics, 69(4), 852-858. https://doi.org/10.1002/prot.21796"
},
{
"DOI": "10.1016/j.bbrc.2004.02.074",
"doi-asserted-by": "crossref",
"key": "75823",
"unstructured": "He, R., Dobie, F., Ballantine, M., Leeson, A., Li, Y., Bastien, N., Cutts, T., Andonov, A., Cao, J., Booth, T. F., Plummer, F. A., Tyler, S., Baker, L. and Li, X. 2004. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochemical and Biophysical Research Communications, 316(2), 476-483. https://doi.org/10.1016/j.bbrc.2004.02.074"
},
{
"DOI": "10.1038/srep32153",
"doi-asserted-by": "crossref",
"key": "75824",
"unstructured": "Heo, L., Lee, H. and Seok, C. 2016. GalaxyRefineComplex: Refinement of protein-protein complex model structures driven by interface repacking. Scientific Reports, 6(1), 32153. https://doi.org/10.1038/srep32153"
},
{
"DOI": "10.1093/nar/gkm259",
"doi-asserted-by": "crossref",
"key": "75825",
"unstructured": "Horton, P., Park, K.J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J. and Nakai, K. 2007. WoLF PSORT: Protein localization predictor. Nucleic Acids Research, 35(suppl_2), W585-W587. https://doi.org/10.1093/nar/gkm259"
},
{
"DOI": "10.1016/j.virusres.2010.08.017",
"doi-asserted-by": "crossref",
"key": "75826",
"unstructured": "Hussain, S. and Gallagher, T. 2010. SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus. Virus Research, 153(2), 299-304. https://doi.org/10.1016/j.virusres.2010.08.017"
},
{
"DOI": "10.22207/JPAM.14.1.03",
"doi-asserted-by": "crossref",
"key": "75827",
"unstructured": "Iqbal, H. M. N., Romero-Castillo, K. D., Bilal, M. and Parra-Saldivar, R. 2020. The emergence of novel-coronavirus and its replication cycle—An overview. Journal of Pure and Applied Microbiology, 14(1). https://doi.org/10.22207/JPAM.14.1.03"
},
{
"DOI": "10.1093/bioinformatics/btt262",
"doi-asserted-by": "crossref",
"key": "75828",
"unstructured": "Jiménez-García, B., Pons, C. and Fernández-Recio, J. 2013. pyDockWEB: A web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics, 29(13), 1698-1699. https://doi.org/10.1093/bioinformatics/btt262"
},
{
"DOI": "10.1038/s41586-021-03819-2",
"doi-asserted-by": "crossref",
"key": "75829",
"unstructured": "Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S. A., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., Back, T., Petersen, S., Reiman, D., Clancy, E., Zielinski, M., Steinegger, M., Pacholska, M., Berghammer, T., Bodenstein, S., Silver, D., Vinyals, O., Senior, A. W., Kavukcuoglu, K., Kohli, P. and Hassabis, D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583-589. https://doi.org/10.1038/s41586-021-03819-2"
},
{
"key": "75830",
"unstructured": "Kannan, S., Shaik Syed Ali, P., Sheeza, A. and Hemalatha, K. 2020. COVID-19 (Novel Coronavirus 2019)—Recent trends. European Review for Medical and Pharmacological Sciences, 24(4), 2006-2011. https://doi.org/10.26355/eurrev_202002_20378"
},
{
"DOI": "10.1016/j.bbrc.2020.11.115",
"doi-asserted-by": "crossref",
"key": "75831",
"unstructured": "Kato, K., Ikliptikawati, D. K., Kobayashi, A., Kondo, H., Lim, K., Hazawa, M. and Wong, R. W. 2021. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochemical and Biophysical Research Communications, 536, 59-66. https://doi.org/10.1016/j.bbrc.2020.11.115"
},
{
"DOI": "10.1093/nar/gkv951",
"doi-asserted-by": "crossref",
"key": "75832",
"unstructured": "Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J. and Bryant, S. H. 2016. PubChem Substance and Compound databases. Nucleic Acids Research, 44(D1), D1202-D1213. https://doi.org/10.1093/nar/gkv951"
},
{
"DOI": "10.1128/JVI.00710-20",
"doi-asserted-by": "crossref",
"key": "75833",
"unstructured": "King, C. R., Tessier, T. M., Dodge, M. J., Weinberg, J. B. and Mymryk, J. S. 2020. Inhibition of Human Adenovirus Replication by the Importin α/β1 Nuclear Import Inhibitor Ivermectin. Journal of Virology, 94(18), e00710-20. https://doi.org/10.1128/JVI.00710-20"
},
{
"DOI": "10.1128/JVI.01782-06",
"doi-asserted-by": "crossref",
"key": "75834",
"unstructured": "Kopecky-Bromberg, S. A., Martínez-Sobrido, L., Frieman, M., Baric, R. A. and Palese, P. 2007. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. Journal of Virology, 81(2), 548-557. https://doi.org/10.1128/JVI.01782-06"
},
{
"DOI": "10.1093/molbev/msy096",
"doi-asserted-by": "crossref",
"key": "75835",
"unstructured": "Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6), 1547-1549. https://doi.org/10.1093/molbev/msy096"
},
{
"DOI": "10.1016/0022-2836(82)90515-0",
"doi-asserted-by": "crossref",
"key": "75836",
"unstructured": "Kyte, J. and Doolittle, R. F. 1982. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology, 157(1), 105-132. https://doi.org/10.1016/0022-2836(82)90515-0"
},
{
"DOI": "10.1107/S0021889892009944",
"doi-asserted-by": "crossref",
"key": "75837",
"unstructured": "Laskowski, R. A., MacArthur, M. W., Moss, D. S. and Thornton, J. M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283-291. https://doi.org/10.1107/S0021889892009944"
},
{
"DOI": "10.1016/j.virusres.2020.198074",
"doi-asserted-by": "crossref",
"key": "75838",
"unstructured": "Li, J.Y., Liao, C.H., Wang, Q., Tan, Y.J., Luo, R., Qiu, Y. and Ge, X.Y. 2020. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Research, 286, 198074. https://doi.org/10.1016/j.virusres.2020.198074"
},
{
"DOI": "10.1093/nar/gkv279",
"doi-asserted-by": "crossref",
"key": "75839",
"unstructured": "Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., Park, Y. M., Buso, N. and Lopez, R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Research, 43(W1), W580-W584. https://doi.org/10.1093/nar/gkv279"
},
{
"DOI": "10.1016/j.antiviral.2014.06.013",
"doi-asserted-by": "crossref",
"key": "75840",
"unstructured": "Liu, D. X., Fung, T. S., Chong, K. K.L., Shukla, A. and Hilgenfeld, R. 2014. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Research, 109, 97-109. https://doi.org/10.1016/j.antiviral.2014.06.013"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"doi-asserted-by": "crossref",
"key": "75841",
"unstructured": "Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., Chen, J., Meng, Y., Wang, J., Lin, Y., Yuan, J., Xie, Z., Ma, J., Liu, W. L., Wang, D., Xu, W., Holmes, E. C., Gao, G. F., Wu, G., Chen, W., Shi, W. and Tan, W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565-574. https://doi.org/10.1016/S0140-6736(20)30251-8"
},
{
"DOI": "10.1093/jac/dks147",
"doi-asserted-by": "crossref",
"key": "75842",
"unstructured": "Mastrangelo, E., Pezzullo, M., De Burghgraeve, T., Kaptein, S., Pastorino, B., Dallmeier, K., de Lamballerie, X., Neyts, J., Hanson, A. M., Frick, D. N., Bolognesi, M. and Milani, M. 2012. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. Journal of Antimicrobial Chemotherapy, 67(8), 1884-1894. https://doi.org/10.1093/jac/dks147"
},
{
"DOI": "10.1038/s41592-022-01488-1",
"doi-asserted-by": "crossref",
"key": "75843",
"unstructured": "Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S. and Steinegger, M. 2022. ColabFold: Making protein folding accessible to all. Nature Methods, 19(6), 679-682. https://doi.org/10.1038/s41592-022-01488-1"
},
{
"DOI": "10.1038/s42003-022-03427-4",
"doi-asserted-by": "crossref",
"key": "75844",
"unstructured": "Miyamoto, Y., Itoh, Y., Suzuki, T., Tanaka, T., Sakai, Y., Koido, M., Hata, C., Wang, C. X., Otani, M., Moriishi, K., Tachibana, T., Kamatani, Y., Yoneda, Y., Okamoto, T. and Oka, M. 2022. SARS-CoV-2 ORF6 disrupts nucleocytoplasmic trafficking to advance viral replication. Communications Biology, 5(1), 1-15. https://doi.org/10.1038/s42003-022-03427-4"
},
{
"DOI": "10.1016/j.virusres.2007.10.009",
"doi-asserted-by": "crossref",
"key": "75845",
"unstructured": "Narayanan, K., Huang, C. and Makino, S. 2008. SARS coronavirus accessory proteins. Virus Research, 133(1), 113-121. https://doi.org/10.1016/j.virusres.2007.10.009"
},
{
"DOI": "10.1016/j.pt.2014.07.005",
"doi-asserted-by": "crossref",
"key": "75846",
"unstructured": "Ōmura, S. and Crump, A. 2014. Ivermectin: Panacea for resource-poor communities? Trends in Parasitology, 30(9), 445-455. https://doi.org/10.1016/j.pt.2014.07.005"
},
{
"DOI": "10.1002/prot.25184",
"doi-asserted-by": "crossref",
"key": "75847",
"unstructured": "Pallara, C., Jiménez-García, B., Romero, M., Moal, I. H. and Fernández-Recio, J. 2017. pyDock scoring for the new modeling challenges in docking: Protein–peptide, homo-multimers, and domain–domain interactions. Proteins: Structure, Function, and Bioinformatics, 85(3), 487-496. https://doi.org/10.1002/prot.25184"
},
{
"DOI": "10.1002/jcc.20084",
"doi-asserted-by": "crossref",
"key": "75848",
"unstructured": "Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. and Ferrin, T. E. 2004. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-1612. https://doi.org/10.1002/jcc.20084"
},
{
"DOI": "10.1016/j.str.2014.11.015",
"doi-asserted-by": "crossref",
"key": "75849",
"unstructured": "Pumroy, R. A., Ke, S., Hart, D. J., Zachariae, U. and Cingolani, G. 2015. Molecular Determinants for Nuclear Import of Influenza A PB2 by Importin α Isoforms 3 and 7. Structure, 23(2), 374-384. https://doi.org/10.1016/j.str.2014.11.015"
},
{
"DOI": "10.1042/BJ20141186",
"doi-asserted-by": "crossref",
"key": "75850",
"unstructured": "Pumroy, R. and Cingolani, G. 2015. Diversification of importin-α isoforms in cellular trafficking and disease states. The Biochemical journal. https://doi.org/10.1042/BJ20141186"
},
{
"DOI": "10.1002/jmv.20095",
"doi-asserted-by": "crossref",
"key": "75851",
"unstructured": "Qinfen, Z., Jinming, C., Xiaojun, H., Huanying, Z., Jicheng, H., Ling, F., Kunpeng, L. and Jingqiang, Z. 2004. The life cycle of SARS coronavirus in Vero E6 cells. Journal of Medical Virology, 73(3), 332-337. https://doi.org/10.1002/jmv.20095"
},
{
"DOI": "10.1007/s00210-020-01902-5",
"doi-asserted-by": "crossref",
"key": "75852",
"unstructured": "Rizzo, E. 2020. Ivermectin, antiviral properties and COVID-19: A possible new mechanism of action. Naunyn-Schmiedeberg’s Archives of Pharmacology, 393(7), 1153-1156. https://doi.org/10.1007/s00210-020-01902-5"
},
{
"DOI": "10.1093/nar/gku316",
"doi-asserted-by": "crossref",
"key": "75853",
"unstructured": "Robert, X. and Gouet, P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(W1), W320-W324. https://doi.org/10.1093/nar/gku316"
},
{
"DOI": "10.1128/JVI.79.17.11507-11512.2005",
"doi-asserted-by": "crossref",
"key": "75854",
"unstructured": "Rowland, R. R. R., Chauhan, V., Fang, Y., Pekosz, A., Kerrigan, M. and Burton, M. D. 2005. Intracellular Localization of the Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein: Absence of Nucleolar Accumulation during Infection and after Expression as a Recombinant Protein in Vero Cells. Journal of Virology, 79(17), 11507-11512. https://doi.org/10.1128/JVI.79.17.11507-11512.2005"
},
{
"DOI": "10.1007/s11224-021-01776-0",
"doi-asserted-by": "crossref",
"key": "75855",
"unstructured": "Saha, J. K. and Raihan, Md. J. 2021. The binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2. Structural Chemistry, 32(5), 1985-1992. https://doi.org/10.1007/s11224-021-01776-0"
},
{
"DOI": "10.1093/emboj/16.23.7067",
"doi-asserted-by": "crossref",
"key": "75856",
"unstructured": "Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T. and Yoneda, Y. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. The EMBO Journal, 16(23), 7067-7077. https://doi.org/10.1093/emboj/16.23.7067"
},
{
"DOI": "10.1080/07391102.2020.1839564",
"doi-asserted-by": "crossref",
"key": "75857",
"unstructured": "Sen Gupta, P. S., Biswal, S., Panda, S. K., Ray, A. K. and Rana, M. K. 2022. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. Journal of Biomolecular Structure and Dynamics, 40(5), 2217-2226. https://doi.org/10.1080/07391102.2020.1839564"
},
{
"DOI": "10.1016/j.jare.2020.03.005",
"doi-asserted-by": "crossref",
"key": "75858",
"unstructured": "Shereen, M. A., Khan, S., Kazmi, A., Bashir, N. and Siddique, R. 2020. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91-98. https://doi.org/10.1016/j.jare.2020.03.005"
},
{
"DOI": "10.1007/978-1-62703-646-7_6",
"doi-asserted-by": "crossref",
"key": "75859",
"unstructured": "Sievers, F. and Higgins, D. G. 2014. Clustal Omega, Accurate Alignment of Very Large Numbers of Sequences. En D. J. Russell (Ed.), Multiple Sequence Alignment Methods (pp. 105-116). Totowa, NJ: Humana Press. https://doi.org/10.1007/978-1-62703-646-7_6"
},
{
"DOI": "10.1042/BJ20040984",
"doi-asserted-by": "crossref",
"key": "75860",
"unstructured": "Surjit, M., Liu, B., Jameel, S., Chow, V. T. K. and Lal, S. K. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochemical Journal, 383(1), 13-18. https://doi.org/10.1042/BJ20040984"
},
{
"DOI": "10.1038/nsmb1212",
"doi-asserted-by": "crossref",
"key": "75861",
"unstructured": "Tarendeau, F., Boudet, J., Guilligay, D., Mas, P. J., Bougault, C. M., Boulo, S., Baudin, F., Ruigrok, R. W. H., Daigle, N., Ellenberg, J., Cusack, S., Simorre, J. P. and Hart, D. J. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Structural & Molecular Biology, 14(3), 229-233. https://doi.org/10.1038/nsmb1212"
},
{
"DOI": "10.1016/j.virusres.2005.05.007",
"doi-asserted-by": "crossref",
"key": "75862",
"unstructured": "Timani, K. A., Liao, Q., Ye, L., Zeng, Y., Liu, J., Zheng, Y., Ye, L., Yang, X., Lingbao, K., Gao, J. and Zhu, Y. 2005. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Research, 114(1), 23-34. https://doi.org/10.1016/j.virusres.2005.05.007"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "crossref",
"key": "75863",
"unstructured": "Trott, O. and Olson, A. J. 2010. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455-461. https://doi.org/10.1002/jcc.21334"
},
{
"DOI": "10.1093/bioinformatics/bty816",
"doi-asserted-by": "crossref",
"key": "75864",
"unstructured": "Vangone, A., Schaarschmidt, J., Koukos, P., Geng, C., Citro, N., Trellet, M. E., Xue, L. C. and Bonvin, A. M. J. J. 2019. Large-scale prediction of binding affinity in protein–small ligand complexes: The PRODIGY-LIG web server. Bioinformatics, 35(9), 1585-1587. https://doi.org/10.1093/bioinformatics/bty816"
},
{
"DOI": "10.1093/nar/gkab1061",
"doi-asserted-by": "crossref",
"key": "75865",
"unstructured": "Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., Yuan, D., Stroe, O., Wood, G., Laydon, A., Žídek, A., Green, T., Tunyasuvunakool, K., Petersen, S., Jumper, J., Clancy, E., Green, R., Vora, A., Lutfi, M., Figurnov, M., Cowie, A., Hobbs, N., Kohli, P., Kleywegt, G., Birney, E., Hassabis, D. and Velankar, S. 2022. AlphaFold Protein Structure Database: Massively expanding the structural coverag2e of protein-sequence space with high-accuracy models. Nucleic Acids Research, 50(D1), D439-D444. https://doi.org/10.1093/nar/gkab1061"
},
{
"DOI": "10.1042/BJ20120150",
"doi-asserted-by": "crossref",
"key": "75866",
"unstructured": "Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D. and Jans, D. A. 2012. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochemical Journal, 443(3), 851-856. https://doi.org/10.1042/BJ20120150"
},
{
"DOI": "10.1002/pro.3330",
"doi-asserted-by": "crossref",
"key": "75867",
"unstructured": "Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall III, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. and Richardson, D. C. 2018. MolProbity: More and better reference data for improved all-atom structure validation. Protein Science, 27(1), 293-315. https://doi.org/10.1002/pro.3330"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "crossref",
"key": "75868",
"unstructured": "Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., Hu, Y., Tao, Z. W., Tian, J. H., Pei, Y. Y., Yuan, M. L., Zhang, Y. L., Dai, F. H., Liu, Y., Wang, Q. M., Zheng, J. J., Xu, L., Holmes, E. C. and Zhang, Y. Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265-269. https://doi.org/10.1038/s41586-020-2008-3"
},
{
"DOI": "10.3389/fmicb.2015.00553",
"doi-asserted-by": "crossref",
"key": "75869",
"unstructured": "Wulan, W. N., Heydet, D., Walker, E. J., Gahan, M. E. and Ghildyal, R. 2015. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Frontiers in Microbiology, 6. https://doi.org/10.3389/fmicb.2015.00553"
},
{
"DOI": "10.1128/JVI.75.19.9345-9356.2001",
"doi-asserted-by": "crossref",
"key": "75870",
"unstructured": "Wurm, T., Chen, H., Hodgson, T., Britton, P., Brooks, G. and Hiscox, J. A. 2001. Localization to the Nucleolus Is a Common Feature of Coronavirus Nucleoproteins, and the Protein May Disrupt Host Cell Division. Journal of Virology, 75(19), 9345-9356. https://doi.org/10.1128/JVI.75.19.9345-9356.2001"
},
{
"DOI": "10.1016/j.chom.2014.07.008",
"doi-asserted-by": "crossref",
"key": "75871",
"unstructured": "Xu, W., Edwards, M. R., Borek, D. M., Feagins, A. R., Mittal, A., Alinger, J. B., Berry, K. N., Yen, B., Hamilton, J., Brett, T. J., Pappu, R. V., Leung, D. W., Basler, C. F. and Amarasinghe, G. K. 2014. Ebola Virus VP24 Targets a Unique NLS Binding Site on Karyopherin Alpha 5 to Selectively Compete with Nuclear Import of Phosphorylated STAT1. Cell Host & Microbe, 16(2), 187-200. https://doi.org/10.1016/j.chom.2014.07.008"
},
{
"DOI": "10.1016/j.antiviral.2020.104760",
"doi-asserted-by": "crossref",
"key": "75872",
"unstructured": "Yang, S. N. Y., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A. and Jans, D. A. 2020. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Research, 177, 104760. https://doi.org/10.1016/j.antiviral.2020.104760"
},
{
"DOI": "10.1016/j.bbagen.2008.07.009",
"doi-asserted-by": "crossref",
"key": "75873",
"unstructured": "Ye, Z., Wong, C. K., Li, P. and Xie, Y. 2008. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochimica et Biophysica Acta (BBA) - General Subjects, 1780(12), 1383-1387. https://doi.org/10.1016/j.bbagen.2008.07.009"
},
{
"DOI": "10.1128/JVI.02371-08",
"doi-asserted-by": "crossref",
"key": "75874",
"unstructured": "Zhao, J., Falcón, A., Zhou, H., Netland, J., Enjuanes, L., Breña, P. P. and Perlman, S. 2009. Severe Acute Respiratory Syndrome Coronavirus Protein 6 Is Required for Optimal Replication. Journal of Virology, 83(5), 2368-2373. https://doi.org/10.1128/JVI.02371-08"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "crossref",
"key": "75875",
"unstructured": "Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J. C., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y. C., Shen, Z. R., Wang, X., Zheng, X. S., Zhao, K., Chen, Q. J., Deng, F., Liu, L. L., Yan, B., Zhan, F. X., Wang, Y. Y., Xiao, G. F. and Shi, Z. L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270-273. https://doi.org/10.1038/s41586-020-2012-7"
}
],
"reference-count": 79,
"references-count": 79,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/view/2485"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach",
"type": "journal-article",
"volume": "27"
}