Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8
et al., Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001, Oct 2024
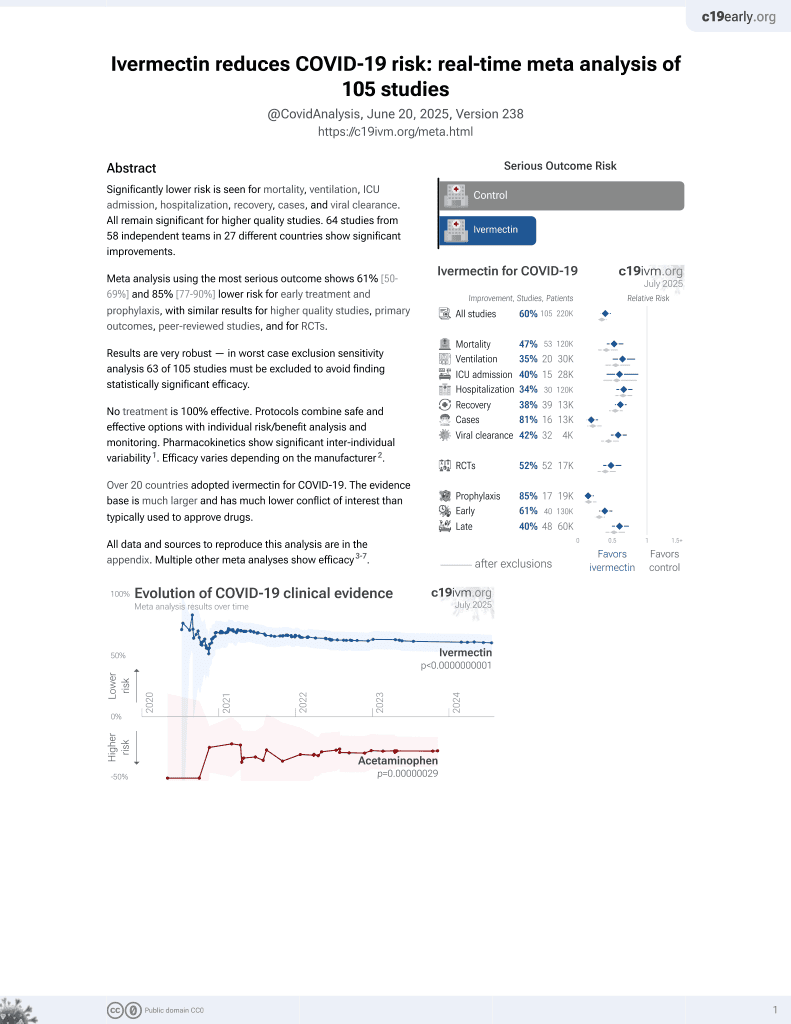
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
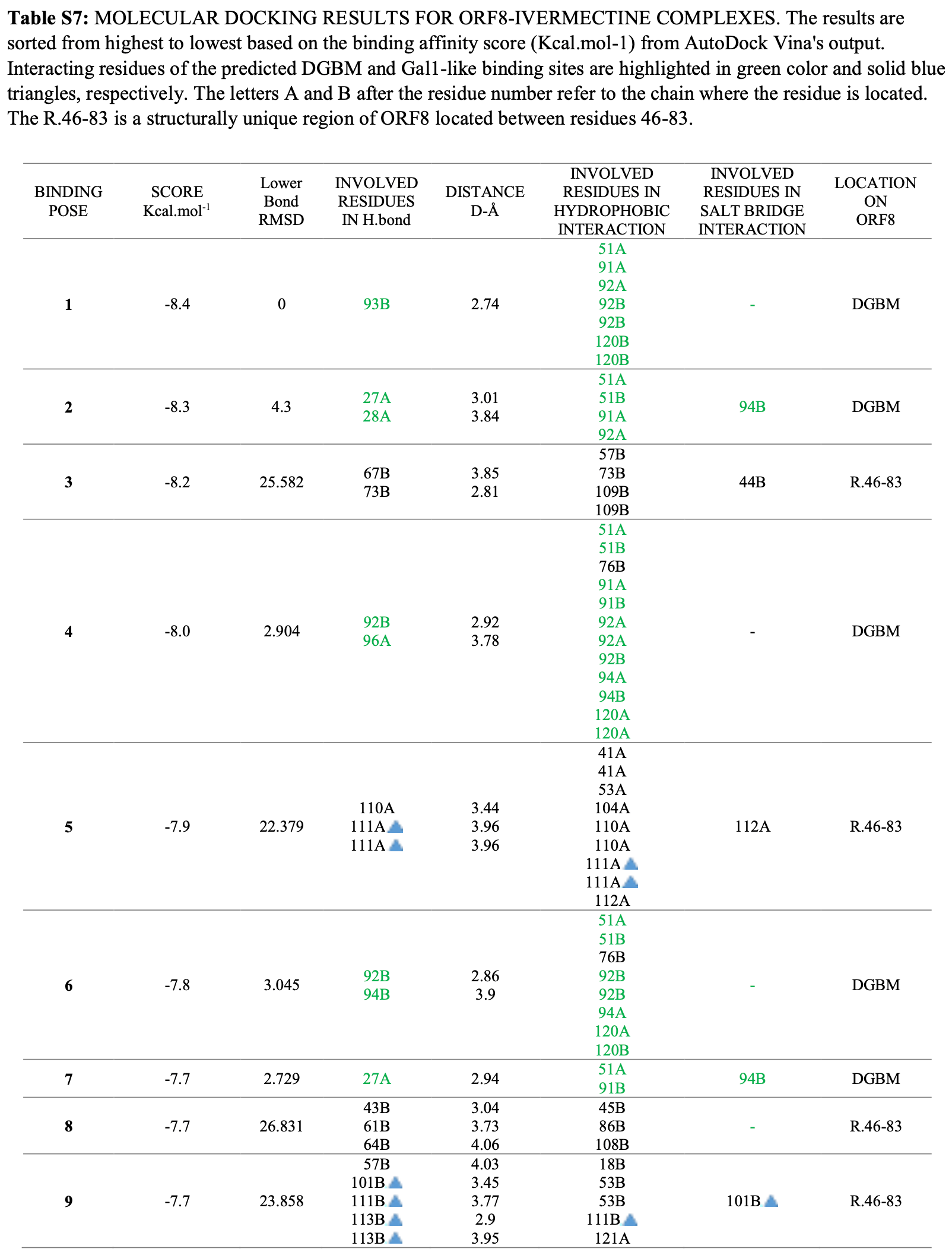
In silico study showing that ivermectin, artemisinin, and DEG-168 may inhibit SARS-CoV-2 by targeting the ORF8 protein's binding sites. Ivermectin showed the highest binding affinity. Authors identified two key binding regions on ORF8 - a deep groove between monomers (DGBM) and a galectin-1-like site. Docking studies found the natural products, especially in combination, effectively bound these sites with high affinity. Inhibiting ORF8 could reduce SARS-CoV-2's immune evasion and pathogenic functions.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
Study covers ivermectin and artemisinin.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Bagheri-Far et al., 26 Oct 2024, peer-reviewed, 5 authors.
Contact: m_azimzadeh@sbu.ac.ir, ma_hosseini@sbu.ac.ir.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8
doi:10.22099/mbrc.2024.50245.2001
The recent pernicious COVID-19 pandemic is caused by SARS-CoV-2. While most therapeutic strategies have focused on the viral spike protein, Open Reading Frame 8 (ORF8) plays a critical role in causing the severity of the disease. Nonetheless, there still needs to be more information on the ORF8 binding epitopes and their appropriate safe inhibitors. Herein, the protein binding sites were detected through comprehensive structural analyses. The validation of the binding sites was investigated through protein conservation analysis and blind docking. The potential natural product (NP) inhibitors were selected based on a structurefunction approach. The solo and combined inhibition functions of these NPs were examined through molecular docking studies. Two binding epitopes were identified, one between the ORF8 monomers (DGBM) and the other on the surface (Gal1-Like). E92 was predicted to be pivotal for DGBM, and R101 for Gal1-like, which was then confirmed through molecular dockings. The inhibitory effects of selected phytochemical (Artemisinin), bacterial (Ivermectin), and native-liken (DEG-168) NPs were compared with the Remdesivir. Selected NPs showed solo-and co-functionality against Remdesivir to inhibit functional regions of the ORF8 structure. The DGBM is highly engaged in capturing the NPs. Additionally, the co-functionality study of NPs showed that the Ivermectin-DEG168 combination has the strongest mechanism for inhibiting all the predicted binding sites. Ivermectin can interfere with ORF8-MHC-I interaction through inhibition of A51 and F120. Two new binding sites on this non-infusion protein structure were introduced using a combination of approaches. Additionally, three safe and effective were found to inhibit these binding sites.
Conflict of Interest: The authors have no relevant financial or non-financial interests to disclose. Authors' Contribution: MBF carried out the analyses and provided all figures and tables and wrote the initial draft. MA re-wrote the manuscript text and citations and revised the manuscript. MAI designed and supervised the project, edited the text, figures and plots, interpreted the data and outlined the manuscript. SMH contributed in interpretation of the results and co-supervised the project. MYA contributed in interpretation of the results.
References
Adasme, Linnemann, Bolz, Kaiser, Salentin et al., PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA, Nucleic Acids Res
Ahmed, Karim, Ross, Hossain, Clemens et al., A fiveday course of ivermectin for the treatment of COVID-19 may reduce the duration of illness, Int J Infect Dis
Alkhansa, Lakkis, El Zein, Mutational analysis of SARS-CoV-2 ORF8 during six months of COVID-19 pandemic, Gene Rep
Assadizadeh, Irani, Oligomer formation of SARS-CoV-2 ORF8 through 73YIDI76 motifs regulates immune response and non-infusion antiviral interactions, Front Mol Biosci
Atanasov, Zotchev, Dirsch, International, Product et al., Natural products in drug discovery: advances and opportunities, Nat Rev Drug Discov
Benson, Cavanaugh, Clark, Karsch-Mizrachi, Lipman et al., GenBank, Nucleic Acids Res
Berman, Battistuz, Bhat, Bluhm, Bourne et al., The Protein Data Bank, Acta Crystallogr D Biol Crystallogr
Biber, Harmelin, Ram, Shaham, Nemet et al., The effect of ivermectin on the viral load and culture viability in early treatment of nonhospitalized patients with mild COVID-19 -a double-blind, randomized placebo-controlled trial, Int J Infect Dis
Bryant, Lawrie, Dowswell, Fordham, Mitchell et al., Ivermectin for Prevention and Treatment of COVID-19 Infection: A Systematic Review, Metaanalysis, and Trial Sequential Analysis to Inform Clinical Guidelines, Am J Ther
Camby, Mercier, Lefranc, Kiss, Galectin-1: a small protein with major functions, Glycobiology
Campbell, History of Avermectin and Ivermectin, with Notes on the History of Other Macrocyclic Lactone Antiparasitic Agents, Curr Pharm Biotechnol
Cavezzi, Troiani, Corrao, COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review, Clin Pract
Chaccour, Hammann, Rabinovich, Ivermectin to reduce malaria transmission I. Pharmacokinetic and pharmacodynamic considerations regarding efficacy and safety, Malar J
Chakravarti, Singh, Ghosh, Dey, Sharma et al., A review on potential of natural products in the management of COVID-19, RSC Adv
Chaudhari, Singh, Joshi, Patel, Joshi, Defective ORF8 dimerization in SARS-CoV-2 delta variant leads to a better adaptive immune response due to abrogation of ORF8-MHC1 interaction, Mol Divers
Chaudhari, Singh, Joshi, Patel, Joshi, Defective ORF8 dimerization in delta variant of SARS CoV2 leads to abrogation of ORF8 MHC-I interaction and overcome suppression of adaptive immune response, bioRxiv
Chen, Zhou, Huang, Zhou, Kang et al., Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses, Front Immunol
Christy, Uekusa, Gerwick, Gerwick, Natural Products with Potential to Treat RNA Virus Pathogens Including SARS-CoV-2, J Nat Prod
Clamp, Cuff, Searle, Barton, The Jalview Java alignment editor, Bioinformatics
Consortium, Pan, Peto, Henao-Restrepo, Preziosi et al., Repurposed Antiviral Drugs for Covid-19 -Interim WHO Solidarity Trial Results, N Engl J Med
Doak, Kihlberg, Drug discovery beyond the rule of 5 -Opportunities and challenges, Expert Opin Drug Discov
Dyshlovenko, Adaptive numerical method for Poisson-Boltzmann equation and its application, Comput Phys Commun
Díaz, SARS-CoV-2 Molecular Network Structure, Front Physiol
Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res
Egbert, Whitty, Keserű, Vajda, Why Some Targets Benefit from beyond Rule of Five Drugs, J Med Chem
Elfiky, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study, Life Sci
Fahmi, Kitagawa, Yasui, Kubota, Ito, The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling, Evol Bioinform Online
Fiser, Sali, ModLoop: automated modeling of loops in protein structures, Bioinformatics
Flower, Buffalo, Hooy, Allaire, Ren et al., Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein, Proc Natl Acad Sci U S A
Fourmont, Revest, Polard, Lederlin, Delaval et al., Acute eosinophilic pneumonia following artenimol-piperaquine exposure, J Travel Med
Gandhi, Klein, Robertson, Peña-Hernández, Lin et al., De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report, Nat Commun
Giguère, Bonin, Cloutier, Patnam, St-Pierre et al., Synthesis of stable and selective inhibitors of human galectins-1 and -3, Bioorg Med Chem
Gong, Tsao, Hsiao, Huang, Huang et al., SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East, Emerg Microbes Infect
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Hachim, Kavian, Cohen, Chin, Chu et al., ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection, Nat Immunol
Hamdorf, Imhof, Bailey-Elkin, Betz, Theobald et al., The unique ORF8 protein from SARS-CoV-2 binds to human dendritic cells and induces a hyper-inflammatory cytokine storm, J Mol Cell Biol
Hetényi, Van Der Spoel, Toward prediction of functional protein pockets using blind docking and pocket search algorithms, Protein Sci
Hsia, Respiratory function of hemoglobin, N Engl J Med
Huey, Morris, Forli, Others, Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial, The Scripps Research Institute Molecular Graphics Laboratory
Humeniuk, Mathias, Kirby, Lutz, Cao et al., Pharmacokinetic, Pharmacodynamic, and Drug-Interaction Profile of Remdesivir, a SARS-CoV-2 Replication Inhibitor, Clin Pharmacokinet
Islam, Parves, Islam, Ali, Efaz et al., Structural and functional effects of the L84S mutant in the SARS-COV-2 ORF8 dimer based on microsecond molecular dynamics study, J Biomol Struct Dyn
Jungreis, Sealfon, Kellis, SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes, Nat Commun
Kandeel, Fayez, Al-Nazawi, From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes, J Med Virol
Kerwin, ChemBioOffice Ultra 2010 Suite, J Am Chem Soc
Kim, Chen, Cheng, Gindulyte, He et al., PubChem 2019 update: improved access to chemical data, Nucleic Acids Res
Krieger, Smeilus, Kaiser, Seo, Efferth et al., Total synthesis and biological investigation of (-)-artemisinin: The antimalarial activity of artemisinin is not stereospecific, Angew Chem Int Ed Engl
Kriplani, Clohisey, Fonseca, Fletcher, Lee et al., Secreted SARS-CoV-2 ORF8 modulates the cytokine expression profile of human macrophages, bioRxiv
Kumar, Nyodu, Maurya, Saxena, Host Immune Response and Immunobiology of Human SARS-CoV-2 Infection, Coronavirus Disease
Lai, Shih, Ko, Tang, Hsueh, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges, Int J Antimicrob Agents
Laing, Devaney, Ivermectin -Old Drug, New Tricks?, Trends Parasitol
Lawrence, Meyerowitz-Katz, Heathers, Brown, Sheldrick, The lesson of ivermectin: meta-analyses based on summary data alone are inherently unreliable, Nat Med
Li, Liao, Wang, Tan, Luo et al., The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway, Virus Res
Li, Weina, Milhous, Pharmacokinetic and Pharmacodynamic Profiles of Rapid-Acting Artemisinins in the Antimalarial Therapy, Curr Drug ther
Lin, Fu, Xiong, Xing, Xue et al., Unconventional secretion of unglycosylated ORF8 is critical for the cytokine storm during SARS-CoV-2 infection, PLoS Pathog
Lin, Fu, Yin, Li, Liu et al., ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway, iScience
Llivisaca-Contreras, Naranjo-Morán, Pino-Acosta, Pieters, Berghe et al., Plants and Natural Products with Activity against Various Types of Coronaviruses: A Review with Focus on SARS-CoV-2, Molecules
Mannan, Ahmed, Arshad, Asim, Qureshi et al., Survey of artemisinin production by diverse Artemisia species in northern Pakistan, Malar J
Matsuoka, Imahashi, Ohno, Ode, Nakata et al., SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer, J Biol Chem
Mcchesney, Natural products in drug discovery--organizing for success, P R Health Sci J
Motlagh, Irani, Nomandan, Assadizadeh, Computational design and investigation of the monomeric spike SARS-CoV-2-ferritin nanocage vaccine stability and interactions, Front Mol Biosci
Nagy, Pongor, Győrffy, Different mutations in SARS-CoV-2 associate with severe and mild outcome, Int J Antimicrob Agents
Nitsure, Sarangi, Shankar, Reddy, Walimbe et al., Mechanisms of Hypoxia in COVID-19 Patients: A Pathophysiologic Reflection, Indian J Crit Care Med
Oostra, De Haan, Rottier, The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8, J Virol
Patridge, Gareiss, Kinch, Hoyer, An analysis of FDA-approved drugs: natural products and their derivatives, Drug Discov Today
Pereira, Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene, Infect Genet Evol
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem
Peña-Silva, Duffull, Steer, Jaramillo-Rincon, Gwee et al., Pharmacokinetic considerations on the repurposing of ivermectin for treatment of COVID-19, Br J Clin Pharmacol
Rashid, Dzakah, Wang, Tang, The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta, Virus Res
Roman, Burela, Pasupuleti, Piscoya, Vidal et al., Ivermectin for the Treatment of Coronavirus Disease 2019: A Systematic Review and Meta-analysis of Randomized Controlled Trials, Clin Infect Dis
Roy, Kucukural, Zhang, I-TASSER: a unified platform for automated protein structure and function prediction, Nat Protoc
Saha, Patgaonkar, Shroff, Ayyar, Bashir et al., Hemoglobin expression in nonerythroid cells: novel or ubiquitous?, Int J Inflam
Salentin, Schreiber, Haupt, Adasme, Schroeder, PLIP: fully automated protein-ligand interaction profiler, Nucleic Acids Res
Schlick, Molecular modeling and simulation: an interdisciplinary guide, doi:10.1007/978-0-387-22464-0
Shandilya, Chacko, Jayaram, Ghosh, A plausible mechanism for the antimalarial activity of artemisinin: A computational approach, Sci Rep
Shoop, Soll, Chemistry, pharmacology and safety of the macrocyclic lactones: ivermectin, abamectin and eprinomectin. Macrocyclic lactones in antiparasitic therapy
Siadat, Direkvand-Moghadam, Study of phytochemical characteristics Artemisia persica Boiss in Ilam Province, Future Natural Products
Sll, The PyMOL molecular graphics system, Version
Sohrabi, Alsafi, Neill, Khan, Kerwan et al., World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19), Int J Surg
St-Pierre, Ouellet, Giguère, Ohtake, Roy et al., Galectin-1specific inhibitors as a new class of compounds to treat HIV-1 infection, Antimicrob Agents Chemother
Su, Young, Linster, Zhu, Jayakumar et al., Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2, MBio
Surade, Blundell, Structural biology and drug discovery of difficult targets: the limits of ligandability, Chem Biol
Tilley, Straimer, Gnädig, Ralph, Fidock, Artemisinin Action and Resistance in Plasmodium falciparum, Trends Parasitol
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem
Valcarcel, Bensussen, Álvarez-Buylla, Díaz, Structural Analysis of SARS-CoV-2 ORF8 Protein: Pathogenic and Therapeutic Implications, Front Genet
Villar, Beglov, Chennamadhavuni, Jr, Kozakov et al., How proteins bind macrocycles, Nat Chem Biol
Vinjamuri, Li, Bouvier, SARS-CoV-2 ORF8: One protein, seemingly one structure, and many functions, Front Immunol
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res
Wang, Wang, Wang, Jiang, Wang et al., SARS-CoV-2 ORF8 Protein Induces Endoplasmic Reticulum Stress-like Responses and Facilitates Virus Replication by Triggering Calnexin: an Unbiased Study, J Virol
Wang, Xu, Wong, Li, Liao et al., Artemisinin, the Magic Drug Discovered from Traditional Chinese Medicine, Proc Est Acad Sci Eng
Waterhouse, Procter, Martin, Clamp, Barton, Jalview Version 2--a multiple sequence alignment editor and analysis workbench, Bioinformatics
Wenzhong, Hualan, COVID-19: ORF8 Synthesizes Nitric Oxide to Break the Blood-Brain/Testi Barrier and Damage the Reproductive System, ChemRxiv
White, Clinical pharmacokinetics and pharmacodynamics of artemisinin and derivatives, Trans R Soc Trop Med Hyg
Wunsch, Mechanical Ventilation in COVID-19: Interpreting the Current Epidemiology, Am J Respir Crit Care Med
Yang, Shen, Hou, Is Ivermectin Effective in Treating COVID-19?, Front Pharmacol
Yang, Zhang, Protein structure and function prediction using I-TASSER, Curr Protoc Bioinformatics
Young, Fong, Chan, Mak, Ang et al., Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study, Lancet
Zhang, Chen, Li, Huang, Luo et al., The ORF8 protein of SARS-CoV-2 mediates immune evasion through downregulating MHC-Ι, Proc Natl Acad Sci
Zhang, I-TASSER server for protein 3D structure prediction, BMC Bioinformatics
Zhou, Gilmore, Ramirez, Settels, Gammeltoft et al., In vitro efficacy of artemisinin-based treatments against SARS-CoV-2, Sci Rep
Ziegel, Berk, Carey, Data Analysis with Microsoft® Excel, Technometrics
DOI record:
{
"DOI": "10.22099/mbrc.2024.50245.2001",
"URL": "https://doi.org/10.22099/mbrc.2024.50245.2001",
"author": [
{
"family": "BagheriFar",
"given": "Mostafa"
},
{
"ORCID": "http://orcid.org/0000-0002-1448-8209",
"family": "Assadizadeh",
"given": "Mohammad"
},
{
"ORCID": "http://orcid.org/0000-0002-1746-8203",
"family": "AzimzadehIrani",
"given": "Maryam"
},
{
"family": "YaghoubiAvini",
"given": "Mohammad"
},
{
"family": "Hosseini",
"given": "Seyed Massoud"
}
],
"container-title": "Molecular Biology Research Communications",
"container-title-short": "Mol Biol Res Commun",
"issue": "Online First",
"issued": {
"date-parts": [
[
2024,
10
]
]
},
"journalAbbreviation": "Mol Biol Res Commun",
"language": "eng",
"publisher": "Shiraz University Press",
"publisher-place": "IR",
"title": "Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8",
"type": "article-journal"
}