In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV
et al., Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2, Mar 2021
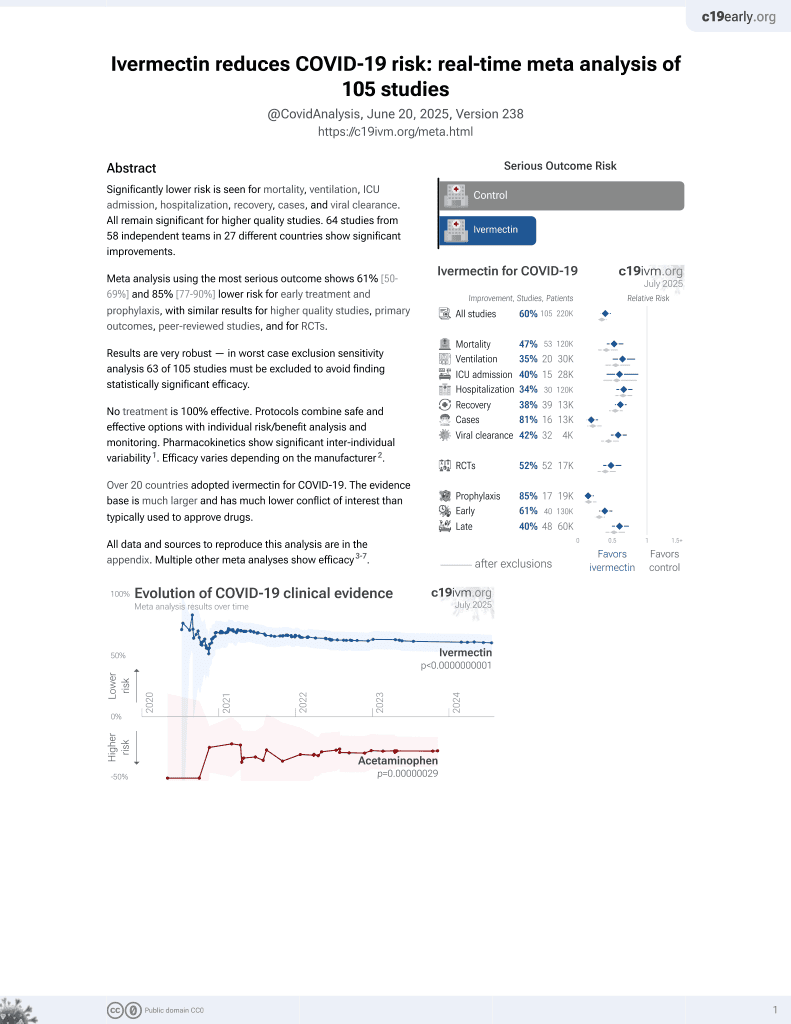
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
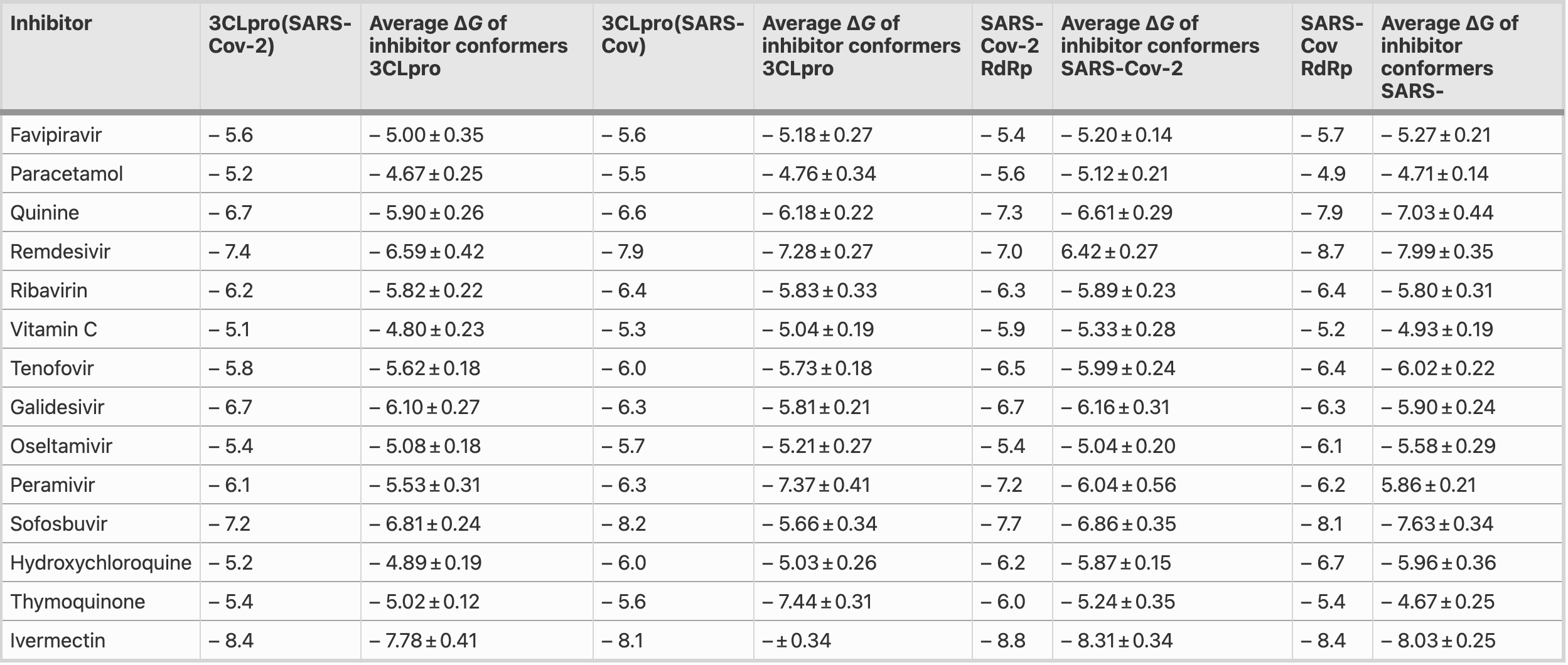
In silico analysis finding that ivermectin had the highest binding energy against the 3CLpro of SARS-CoV-2 and RdRps of both SARS-CoV and SARS-CoV-2.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Udofia et al., 25 Mar 2021, peer-reviewed, 5 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV
Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2
An outbreak of a cluster of viral pneumonia cases, subsequently identified as coronavirus disease 2019 , due to a novel SARS-CoV-2 necessitates an urgent need for a vaccine to prevent infection or an approved medication for a cure. In our in silico molecular docking study, a total of 173 compounds, including FDA-approved antiviral drugs, with good ADME descriptors, and some other nucleotide analogues were screened. The results show that these compounds demonstrate strong binding affinity for the residues at the active sites of RNA-dependent RNA-polymerase (RdRp) modelled structures and Chymotrypsin-like cysteine protease (3CLpro) of the HCoV proteins. Free energies (ΔG's) of binding for SARS-CoV-2 and SARS-CoV RdRp range from -5.4 to -8.8 kcal/mol and -4.9 to -8.7 kcal/mol, respectively. Also, SARS-CoV-2 and SARS-CoV 3CLpro gave ΔG values ranging from − 5.1 to − 8.4 kcal/mol and − 5.5 to − 8.6 kcal/mol, respectively. Interesting results are obtained for ivermectin, an antiparasitic agent with broad spectrum activity, which gave the highest binding energy value (− 8.8 kcal/mol) against the 3CLpro of SARS-CoV-2 and RdRps of both SARS-CoV and SARS-CoV-2. The reason for such high binding energy values is probably due to the presence of hydroxy, methoxy and sugar moieties in its structure. The stability of the protein-ligand complexes of polymerase inhibitors considered in this investigation, such as Sofosbuvir, Remdesivir, Tenofovir, Ribavirin, Galidesivir, 5c3, 5h1 and 7a1, show strong to moderate hydrogen bonding and hydrophobic interactions (π-π stacked, π-π T-shaped, π-sigma and π-alkyl). The stability provided from such interactions translate into greater antiviral activity or inhibitory effect of the ligands. Assessment of the average free energies of binding of the FDA approved drugs are highly comparable for conformers of a particular inhibitor, indicating similar modes of binding within the pockets.
Supplementary Information The online version contains supplementary material available at https:// doi. org/ 10. 1007/ s13721-021-00299-2.
References
Andersen, Rambaut, Lipkin, Holmes, Garry, The proximal origin of SARS-CoV-2, Nat Med, doi:10.1038/s41591-020-0820-9
Bajji, Davis, Synthesis and biophysical characterization of tRNALys,3 anticodon stem-loop RNAs containing the mcm5s2U nucleoside, Org Lett, doi:10.1021/ol006605h
Beg, Shivangi, Meena, Structural Prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H 37 Rv to determine its essentiality in survival, Adv Bioinform, doi:10.1155/2018/6152014
Ben-Zvi, Kivity, Langevitz, Shoenfeld, Hydroxychloroquine: from malaria to autoimmunity, Clin Rev Allergy Immunol
Biasini, Bienert, Waterhouse, Arnold, Studer et al., SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information, Nucleic Acids Res, doi:10.1093/nar/gku340
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antivir Res, doi:10.1016/j.antiviral.2020.104787
Cao, Xiao, Cao, Li, Kumaki et al., Inhibition of novel reassortant avian influenza H7N9 virus infection in vitro with three antiviral drugs, oseltamivir, peramivir and favipiravir, Antiviral Chem Chemother, doi:10.3851/IMP2672
Chaccour, Hammann, Ramón-García, Rabinovich, Ivermectin and Novel Coronavirus Disease (COVID-19): Keeping Rigor in Times of Urgency, Am J Trop Med Hygiene, doi:10.4269/ajtmh.20-0271
Chan, Chan, Tracing the SARS-coronavirus, J Thorac Dis, doi:10.3978/j.issn.2072-1439.2013.06.19
Chen, Yiu, Wong, Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates, F1000Research, doi:10.12688/f1000research.22457.1
Cheng, Zhang, Xie, Jiang, Arnold et al., Expression, purification, and characterization of SARS coronavirus RNA polymerase, Virology, doi:10.1016/j.virol.2005.02.017
Colson, Rolain, Lagier, Brouqui, Raoult, Chloroquine and hydroxychloroquine as available weapons to fight COVID-19, Int J Antimicrob Agents, doi:10.1016/j.ijantimicag.2020.105932
Corman, Muth, Niemeyer, Drosten, Hosts and sources of endemic human coronaviruses, Advances in virus research, doi:10.1016/bs.aivir.2018.01.001
Dallakyan, Olson, Small-molecule library screening by docking with PyRx BT-chemical biology: methods and protocols, doi:10.1007/978-1-4939-2269-7_19
Erion, Bullough, Lin, Hong, HepDirect prodrugs for targeting nucleotide-based antiviral drugs to the liver, Curr Opin Investig Drugs
Fantini, Scala, Chahinian, Yahi, Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection, Int J Antimicrob Agents, doi:10.1016/j.ijantimicag.2020.105960
Flierl, Nero, Lim, Arthur, Yao et al., Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators, J Exp Med, doi:10.1084/jem.20140391
Fox, Dixon, Guarrasi, Krubel, Treatment of primary Sjögren's syndrome with hydroxychloroquine: a retrospective, open-label study, Lupus
Fu, Li, Zhang, Strong orbital Interaction in pi-pi Stacking System
Gao, Gao, Yan, Huang, Liu et al., Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science
Gao, Tian, Yang, Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends, doi:10.5582/bst.2020.01047
Hasan, Hossain, Analysis of COVID-19 M protein for possible clues regarding virion stability , longevity and spreading, doi:10.31219/osf.io/e7jkc
Henderleiter, Smart, Anderson, Elian, How do organic chemistry students understand and apply hydrogen bonding?, J Chem Educ, doi:10.1021/ed078p1126
Huang, Bosch, Li, Li, Kyoung et al., SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells, J Biol Chem, doi:10.1074/jbc.M508381200
Jeffrey, An introduction to hydrogen bonding
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Ju, Kumar, Li, Jockusch, Russo, Nucleotide analogues as inhibitors of viral polymerases, BioRxiv, doi:10.1101/2020.01.30.927574
Ju, Li, Kumar, Jockusch, Chien et al., Nucleotide Analogues as Inhibitors of SARS-CoV Polymerase, BioRxiv, doi:10.1101/2020.03.12.989186
Khan, Zia, Ashraf, Uddin, Ul-Haq, Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1751298
Kirchdoerfer, Ward, Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors, Nat Commun, doi:10.1038/s41467-019-10280-3
Lee, Yang, Parr, Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys Rev B, doi:10.1103/PhysRevB.37.785
Li, Clercq, Therapeutic options for the 2019 novel coronavirus (2019-nCoV), Nat Rev Drug Discov, doi:10.1038/d41573-020-00016-0
Li, Moore, Vasilieva, Sui, Wong et al., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus, Nature, doi:10.1038/nature02145
Lüthy, Bowie, Eisenberg, Assessment of protein models with three-dimensional profiles, Nature, doi:10.1038/356083a0
Martin, Canard, Decroly, Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle, Antivir Res, doi:10.1016/j.antiviral.2017.02.004
Martinez, Iverson, Rethinking the term "pi-stacking, Chem Sci, doi:10.1039/C2SC20045G
Masters, The molecular biology of coronaviruses, Adv Virus Res, doi:10.1016/S0065-3527(06)66005-3
Pamidighantam, Nakandala, Abeysinghe, Wimalasena, Yodage et al., Community science exemplars in SEAGrid science gateway: apache airavata based implementation of advanced infrastructure, Proc Comput Sci, doi:10.1016/j.procs.2016.05.535
Panigrahi, Strong and weak hydrogen bonds in proteinligand complexes of kinases: a comparative study, Amino Acids, doi:10.1007/s00726-007-0015-4
Patil, Balasubramanian, Masand, Chapter 14-dengue virus polymerase: a crucial target for antiviral drug discovery, doi:10.1016/B978-0-12-815422-9.00014-0
Pearson, Using the FASTA program to search protein and DNA sequence databases, doi:10.1385/0-89603-246-9:307
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera-a visualization system for exploratory research and analysis, J Comput Chem
Prabakaran, Xiao, Dimitrov, A model of the ACE2 structure and function as a SARS-CoV receptor, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2003.12.081
Raha, Merz, Chapter 9 Calculating binding free energy in protein-ligand interaction, Annual reports in computational chemistry, doi:10.1016/S1574-1400(05)01009-1
Shi, Sivaraman, Song, Mechanism for controlling the dimermonomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease, J Virol, doi:10.1128/JVI.02680-07
Solowiej, Thomson, Ryan, Luo, He et al., Steady-State and pre-steady-state kinetic evaluation of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro Cysteine protease: development of an ion-pair model for catalysis, Biochemistry, doi:10.1021/bi702107v
Spaan, Cavanagh, Horzinek, Coronaviruses: structure and genome expression, J Gen Virol, doi:10.1099/0022-1317-69-12-2939
Spiwok, CH/π Interactions in Carbohydrate Recognition, Molecules, doi:10.3390/molecules22071038
Tomar, Mudgal, Pareek, Chapter 3-RNA-dependent RNA polymerase of alphaviruses: a potential target for the design of drugs against alphaviruses, doi:10.1016/B978-0-12-815422-9.00003-6
Touret, De Lamballerie, Of chloroquine and COVID-19, Antivir Res, doi:10.1016/j.antiviral.2020.104762
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem, doi:10.1002/jcc.21334
Van Doremalen, Bushmaker, Morris, Holbrook, Gamble et al., Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1, N Engl J Med, doi:10.1056/NEJMc2004973
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, doi:10.1016/j.cell.2020.02.058
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res, doi:10.1038/s41422-020-0282-0
Wang, Lai, Wang, Further development and validation of empirical scoring functions for structure-based binding affinity 22 Page 12 of 12 prediction, J Comput Aided Mol Des, doi:10.1023/A:1016357811882
Wang, Wang, Chen, Qin, Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures, J Med Virol, doi:10.1002/jmv.25748
Waterhouse, Bertoni, Bienert, Studer, Tauriello et al., SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res, doi:10.1093/nar/gky427
Who, Coronavirus disease (COVID, doi:10.1093/nar/gkz966
Williams, Headd, Moriarty, Prisant, Videau et al., MolProbity: More and better reference data for improved all-atom structure validation, Protein Sci, doi:10.1002/pro.3330
Yang, Roy, Zhang, Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment, Bioinformatics, doi:10.1093/bioinformatics/btt447
Yang, Yang, Ding, Liu, Lou et al., The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor, Proc Natl Acad Sci
DOI record:
{
"DOI": "10.1007/s13721-021-00299-2",
"ISSN": [
"2192-6662",
"2192-6670"
],
"URL": "http://dx.doi.org/10.1007/s13721-021-00299-2",
"alternative-id": [
"299"
],
"article-number": "22",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "31 October 2020"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Revised",
"name": "revised",
"order": 2,
"value": "25 January 2021"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 3,
"value": "12 March 2021"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 4,
"value": "25 March 2021"
},
{
"label": "Free to read",
"name": "free",
"value": "This content has been made available to all."
}
],
"author": [
{
"affiliation": [],
"family": "Udofia",
"given": "Inemesit A.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Gbayo",
"given": "Kofoworola O.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Oloba-Whenu",
"given": "Oluwakemi A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ogunbayo",
"given": "Taofeek B.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6633-6066",
"affiliation": [],
"authenticated-orcid": false,
"family": "Isanbor",
"given": "Chukwuemeka",
"sequence": "additional"
}
],
"container-title": "Network Modeling Analysis in Health Informatics and Bioinformatics",
"container-title-short": "Netw Model Anal Health Inform Bioinforma",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2021,
3,
25
]
],
"date-time": "2021-03-25T21:59:06Z",
"timestamp": 1616709546000
},
"deposited": {
"date-parts": [
[
2021,
12,
9
]
],
"date-time": "2021-12-09T08:21:50Z",
"timestamp": 1639038110000
},
"indexed": {
"date-parts": [
[
2023,
6,
27
]
],
"date-time": "2023-06-27T19:12:35Z",
"timestamp": 1687893155569
},
"is-referenced-by-count": 5,
"issue": "1",
"issued": {
"date-parts": [
[
2021,
3,
25
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2021,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.springer.com/tdm",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
3,
25
]
],
"date-time": "2021-03-25T00:00:00Z",
"timestamp": 1616630400000
}
},
{
"URL": "https://www.springer.com/tdm",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
3,
25
]
],
"date-time": "2021-03-25T00:00:00Z",
"timestamp": 1616630400000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s13721-021-00299-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s13721-021-00299-2/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s13721-021-00299-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1007",
"published": {
"date-parts": [
[
2021,
3,
25
]
]
},
"published-online": {
"date-parts": [
[
2021,
3,
25
]
]
},
"published-print": {
"date-parts": [
[
2021,
12
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1038/s41591-020-0820-9",
"author": "KG Andersen",
"doi-asserted-by": "publisher",
"first-page": "44",
"issue": "1",
"journal-title": "Nat Med",
"key": "299_CR1",
"unstructured": "Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF (2020) The proximal origin of SARS-CoV-2. Nat Med 89(1):44–48. https://doi.org/10.1038/s41591-020-0820-9",
"volume": "89",
"year": "2020"
},
{
"DOI": "10.1021/ol006605h",
"author": "AC Bajji",
"doi-asserted-by": "publisher",
"first-page": "3865",
"issue": "24",
"journal-title": "Org Lett",
"key": "299_CR2",
"unstructured": "Bajji AC, Davis DR (2000) Synthesis and biophysical characterization of tRNALys,3 anticodon stem-loop RNAs containing the mcm5s2U nucleoside. Org Lett 2(24):3865–3868. https://doi.org/10.1021/ol006605h",
"volume": "2",
"year": "2000"
},
{
"DOI": "10.1155/2018/6152014",
"author": "MA Beg",
"doi-asserted-by": "publisher",
"journal-title": "Adv Bioinform",
"key": "299_CR3",
"unstructured": "Beg MA, Shivangi TSC, Meena LS (2018) Structural Prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Adv Bioinform. https://doi.org/10.1155/2018/6152014",
"year": "2018"
},
{
"DOI": "10.1007/s12016-010-8243-x",
"author": "I Ben-Zvi",
"doi-asserted-by": "publisher",
"first-page": "145",
"issue": "2",
"journal-title": "Clin Rev Allergy Immunol",
"key": "299_CR01",
"unstructured": "Ben-Zvi I, Kivity S, Langevitz P, Shoenfeld Y (2012) Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol 42(2):145–153",
"volume": "42",
"year": "2012"
},
{
"DOI": "10.1093/nar/gku340",
"author": "M Biasini",
"doi-asserted-by": "publisher",
"first-page": "W252",
"issue": "W1",
"journal-title": "Nucleic Acids Res",
"key": "299_CR4",
"unstructured": "Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42(W1):W252–W258. https://doi.org/10.1093/nar/gku340",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"author": "L Caly",
"doi-asserted-by": "publisher",
"journal-title": "Antivir Res",
"key": "299_CR5",
"unstructured": "Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res. https://doi.org/10.1016/j.antiviral.2020.104787",
"year": "2020"
},
{
"DOI": "10.3851/IMP2672",
"author": "R-Y Cao",
"doi-asserted-by": "publisher",
"first-page": "237",
"issue": "6",
"journal-title": "Antiviral Chem Chemother",
"key": "299_CR6",
"unstructured": "Cao R-Y, Xiao J-H, Cao B, Li S, Kumaki Y, Zhong W (2014) Inhibition of novel reassortant avian influenza H7N9 virus infection in vitro with three antiviral drugs, oseltamivir, peramivir and favipiravir. Antiviral Chem Chemother 23(6):237–240. https://doi.org/10.3851/IMP2672",
"volume": "23",
"year": "2014"
},
{
"DOI": "10.4269/ajtmh.20-0271",
"author": "C Chaccour",
"doi-asserted-by": "publisher",
"journal-title": "Am J Trop Med Hygiene",
"key": "299_CR7",
"unstructured": "Chaccour C, Hammann F, Ramón-García S, Rabinovich NR (2020) Ivermectin and Novel Coronavirus Disease (COVID-19): Keeping Rigor in Times of Urgency. Am J Trop Med Hygiene. https://doi.org/10.4269/ajtmh.20-0271",
"year": "2020"
},
{
"DOI": "10.3978/j.issn.2072-1439.2013.06.19",
"author": "PKS Chan",
"doi-asserted-by": "publisher",
"first-page": "S118",
"issue": "Suppl 2",
"journal-title": "J Thorac Dis",
"key": "299_CR8",
"unstructured": "Chan PKS, Chan MCW (2013) Tracing the SARS-coronavirus. J Thorac Dis 5 Suppl 2(Suppl 2):S118–S121. https://doi.org/10.3978/j.issn.2072-1439.2013.06.19",
"volume": "5 Suppl 2",
"year": "2013"
},
{
"DOI": "10.12688/f1000research.22457.1",
"author": "YW Chen",
"doi-asserted-by": "publisher",
"first-page": "129",
"journal-title": "F1000Research",
"key": "299_CR9",
"unstructured": "Chen YW, Yiu C-PB, Wong K-Y (2020) Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 9:129. https://doi.org/10.12688/f1000research.22457.1",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1016/j.virol.2005.02.017",
"author": "A Cheng",
"doi-asserted-by": "publisher",
"first-page": "165",
"issue": "2",
"journal-title": "Virology",
"key": "299_CR10",
"unstructured": "Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, Sarafianos SG, Ding J (2005) Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335(2):165–176. https://doi.org/10.1016/j.virol.2005.02.017",
"volume": "335",
"year": "2005"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105932",
"author": "P Colson",
"doi-asserted-by": "publisher",
"journal-title": "Int J Antimicrob Agents",
"key": "299_CR11",
"unstructured": "Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D (2020) Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. https://doi.org/10.1016/j.ijantimicag.2020.105932",
"year": "2020"
},
{
"DOI": "10.1016/bs.aivir.2018.01.001",
"doi-asserted-by": "publisher",
"key": "299_CR12",
"unstructured": "Corman VM, Muth D, Niemeyer D, Drosten C (2018) Hosts and sources of endemic human coronaviruses. In: Advances in virus research, vol. 100. Elsevier Inc., pp 163–188. https://doi.org/10.1016/bs.aivir.2018.01.001"
},
{
"DOI": "10.1007/978-1-4939-2269-7_19",
"author": "S Dallakyan",
"doi-asserted-by": "publisher",
"first-page": "243",
"key": "299_CR13",
"unstructured": "Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx BT—chemical biology: methods and protocols. In: Hempel JE, Williams CH, Hong CC (eds) Chemical biology. Springer New York, pp 243–250. https://doi.org/10.1007/978-1-4939-2269-7_19",
"volume-title": "Chemical biology",
"year": "2015"
},
{
"key": "299_CR14",
"unstructured": "Erion MD, Bullough DA, Lin C-C, Hong Z (2006). HepDirect prodrugs for targeting nucleotide-based antiviral drugs to the liver. Curr Opin Investig Drugs (London, England : 2000) 7(2): 109–117. http://europepmc.org/abstract/MED/16499280. Accessed 23 Apr 2020"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105960",
"author": "J Fantini",
"doi-asserted-by": "publisher",
"journal-title": "Int J Antimicrob Agents",
"key": "299_CR15",
"unstructured": "Fantini J, Di Scala C, Chahinian H, Yahi N (2020) Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. https://doi.org/10.1016/j.ijantimicag.2020.105960",
"year": "2020"
},
{
"DOI": "10.1084/jem.20140391",
"author": "U Flierl",
"doi-asserted-by": "publisher",
"first-page": "129",
"issue": "2",
"journal-title": "J Exp Med",
"key": "299_CR16",
"unstructured": "Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MTK, Jandrot-Perrus M, Schäfer A, Nieswandt B, Andrews RK, Parker MW, Gardiner EE, Peter K (2015) Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators. J Exp Med 212(2):129–137. https://doi.org/10.1084/jem.20140391",
"volume": "212",
"year": "2015"
},
{
"DOI": "10.1177/0961203396005001081",
"author": "RI Fox",
"doi-asserted-by": "publisher",
"first-page": "31",
"issue": "1_suppl",
"journal-title": "Lupus",
"key": "299_CR02",
"unstructured": "Fox RI, Dixon R, Guarrasi V, Krubel S (1996) Treatment of primary Sjögren's syndrome with hydroxychloroquine: a retrospective, open-label study. Lupus 5(1_suppl):31–36",
"volume": "5",
"year": "1996"
},
{
"key": "299_CR17",
"unstructured": "Fu X-X, Li J-F, Zhang R-Q (2016) Strong orbital Interaction in pi-pi Stacking System. In: arXiv e-prints arXiv:1601.01150. https://ui.adsabs.harvard.edu/abs/2016arXiv160101150F. Accessed 23 Apr 2020"
},
{
"DOI": "10.5582/bst.2020.01047",
"author": "J Gao",
"doi-asserted-by": "publisher",
"first-page": "72",
"issue": "1",
"journal-title": "Biosci Trends",
"key": "299_CR18",
"unstructured": "Gao J, Tian Z, Yang X (2020a) Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14(1):72–73. https://doi.org/10.5582/bst.2020.01047",
"volume": "14",
"year": "2020"
},
{
"DOI": "10.1126/science.abb7498",
"author": "Y Gao",
"doi-asserted-by": "publisher",
"first-page": "779",
"issue": "6492",
"journal-title": "Science",
"key": "299_CR19",
"unstructured": "Gao Y, Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Wang Q, Lou Z, Rao Z (2020b) Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368(6492):779–782",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.31219/osf.io/e7jkc",
"doi-asserted-by": "publisher",
"key": "299_CR20",
"unstructured": "Hasan S, Hossain MM (2020) Analysis of COVID-19 M protein for possible clues regarding virion stability , longevity and spreading. https://doi.org/10.31219/osf.io/e7jkc"
},
{
"DOI": "10.1021/ed078p1126",
"author": "J Henderleiter",
"doi-asserted-by": "publisher",
"first-page": "1126",
"issue": "8",
"journal-title": "J Chem Educ",
"key": "299_CR21",
"unstructured": "Henderleiter J, Smart R, Anderson J, Elian O (2001) How do organic chemistry students understand and apply hydrogen bonding? J Chem Educ 78(8):1126. https://doi.org/10.1021/ed078p1126",
"volume": "78",
"year": "2001"
},
{
"DOI": "10.1074/jbc.M508381200",
"author": "IC Huang",
"doi-asserted-by": "publisher",
"first-page": "3198",
"issue": "6",
"journal-title": "J Biol Chem",
"key": "299_CR22",
"unstructured": "Huang IC, Bosch BJ, Li F, Li W, Kyoung HL, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, Farzan M, Rottier PJM, Choe H (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 281(6):3198–3203. https://doi.org/10.1074/jbc.M508381200",
"volume": "281",
"year": "2006"
},
{
"key": "299_CR23",
"unstructured": "Jeffrey GA (1997) An introduction to hydrogen bonding (12th ed.). Oxford University Press. https://books.google.fr/books?id=ZRAFifo37QsC. Accessed 23 Apr 2020"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"author": "Z Jin",
"doi-asserted-by": "publisher",
"first-page": "289",
"issue": "7811",
"journal-title": "Nature",
"key": "299_CR24",
"unstructured": "Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang H (2020) Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582(7811):289–293. https://doi.org/10.1038/s41586-020-2223-y",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1101/2020.01.30.927574",
"author": "J Ju",
"doi-asserted-by": "publisher",
"journal-title": "BioRxiv",
"key": "299_CR25",
"unstructured": "Ju J, Kumar S, Li X, Jockusch S, Russo JJ (2020a) Nucleotide analogues as inhibitors of viral polymerases. BioRxiv. https://doi.org/10.1101/2020.01.30.927574",
"year": "2020"
},
{
"DOI": "10.1101/2020.03.12.989186",
"author": "J Ju",
"doi-asserted-by": "publisher",
"journal-title": "BioRxiv",
"key": "299_CR26",
"unstructured": "Ju J, Li X, Kumar S, Jockusch S, Chien M, Tao C, Morozova I, Kalachikov S, Kirchdoerfer RN, Russo JJ (2020b) Nucleotide Analogues as Inhibitors of SARS-CoV Polymerase. BioRxiv. https://doi.org/10.1101/2020.03.12.989186",
"year": "2020"
},
{
"DOI": "10.1080/07391102.2020.1751298",
"author": "SA Khan",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "J Biomol Struct Dyn",
"key": "299_CR27",
"unstructured": "Khan SA, Zia K, Ashraf S, Uddin R, Ul-Haq Z (2020) Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn 9(1):1–10. https://doi.org/10.1080/07391102.2020.1751298",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1038/s41467-019-10280-3",
"author": "RN Kirchdoerfer",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Nat Commun",
"key": "299_CR28",
"unstructured": "Kirchdoerfer RN, Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10(1):1–9. https://doi.org/10.1038/s41467-019-10280-3",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1103/PhysRevB.37.785",
"author": "C Lee",
"doi-asserted-by": "publisher",
"first-page": "785",
"issue": "2",
"journal-title": "Phys Rev B",
"key": "299_CR29",
"unstructured": "Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37(2):785–789. https://doi.org/10.1103/PhysRevB.37.785",
"volume": "37",
"year": "1988"
},
{
"DOI": "10.1038/d41573-020-00016-0",
"author": "G Li",
"doi-asserted-by": "publisher",
"first-page": "149",
"issue": "3",
"journal-title": "Nat Rev Drug Discov",
"key": "299_CR30",
"unstructured": "Li G, De Clercq E (2020) Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 19(3):149–150. https://doi.org/10.1038/d41573-020-00016-0",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1038/nature02145",
"author": "W Li",
"doi-asserted-by": "publisher",
"first-page": "450",
"issue": "6965",
"journal-title": "Nature",
"key": "299_CR31",
"unstructured": "Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426(6965):450–454. https://doi.org/10.1038/nature02145",
"volume": "426",
"year": "2003"
},
{
"DOI": "10.1038/356083a0",
"author": "R Lüthy",
"doi-asserted-by": "publisher",
"first-page": "83",
"issue": "6364",
"journal-title": "Nature",
"key": "299_CR32",
"unstructured": "Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83–85. https://doi.org/10.1038/356083a0",
"volume": "356",
"year": "1992"
},
{
"DOI": "10.1016/j.antiviral.2017.02.004",
"author": "B Martin",
"doi-asserted-by": "publisher",
"first-page": "48",
"journal-title": "Antivir Res",
"key": "299_CR33",
"unstructured": "Martin B, Canard B, Decroly E (2017) Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle. Antivir Res 141:48–61. https://doi.org/10.1016/j.antiviral.2017.02.004",
"volume": "141",
"year": "2017"
},
{
"DOI": "10.1039/C2SC20045G",
"author": "CR Martinez",
"doi-asserted-by": "publisher",
"first-page": "2191",
"issue": "7",
"journal-title": "Chem Sci",
"key": "299_CR34",
"unstructured": "Martinez CR, Iverson BL (2012) Rethinking the term “pi-stacking.” Chem Sci 3(7):2191–2201. https://doi.org/10.1039/C2SC20045G",
"volume": "3",
"year": "2012"
},
{
"DOI": "10.1016/S0065-3527(06)66005-3",
"author": "PS Masters",
"doi-asserted-by": "publisher",
"first-page": "193",
"issue": "06",
"journal-title": "Adv Virus Res",
"key": "299_CR35",
"unstructured": "Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res 65(06):193–292. https://doi.org/10.1016/S0065-3527(06)66005-3",
"volume": "65",
"year": "2006"
},
{
"DOI": "10.1016/j.procs.2016.05.535",
"author": "S Pamidighantam",
"doi-asserted-by": "publisher",
"first-page": "1927",
"journal-title": "Proc Comput Sci",
"key": "299_CR36",
"unstructured": "Pamidighantam S, Nakandala S, Abeysinghe E, Wimalasena C, Yodage SR, Marru S, Pierce M (2016) Community science exemplars in SEAGrid science gateway: apache airavata based implementation of advanced infrastructure. Proc Comput Sci 80:1927–1939. https://doi.org/10.1016/j.procs.2016.05.535",
"volume": "80",
"year": "2016"
},
{
"DOI": "10.1007/s00726-007-0015-4",
"author": "SK Panigrahi",
"doi-asserted-by": "publisher",
"first-page": "617",
"issue": "4",
"journal-title": "Amino Acids",
"key": "299_CR37",
"unstructured": "Panigrahi SK (2008) Strong and weak hydrogen bonds in protein-ligand complexes of kinases: a comparative study. Amino Acids 34(4):617–633. https://doi.org/10.1007/s00726-007-0015-4",
"volume": "34",
"year": "2008"
},
{
"DOI": "10.1016/B978-0-12-815422-9.00014-0",
"author": "VM Patil",
"doi-asserted-by": "publisher",
"first-page": "387",
"key": "299_CR38",
"unstructured": "Patil VM, Balasubramanian K, Masand N (2019) Chapter 14—dengue virus polymerase: a crucial target for antiviral drug discovery. In: Gupta SPBT-VP (ed) Viral polymerases. Academic Press, pp 387–428. https://doi.org/10.1016/B978-0-12-815422-9.00014-0",
"volume-title": "Viral polymerases",
"year": "2019"
},
{
"DOI": "10.1385/0-89603-246-9:307",
"author": "WR Pearson",
"doi-asserted-by": "publisher",
"first-page": "307",
"key": "299_CR39",
"unstructured": "Pearson WR (1994) Using the FASTA program to search protein and DNA sequence databases. In: Griffin AM, Griffin HG (eds) Computer analysis of sequence data. Humana Press, pp 307–331. https://doi.org/10.1385/0-89603-246-9:307",
"volume-title": "Computer analysis of sequence data",
"year": "1994"
},
{
"DOI": "10.1002/jcc.20084",
"author": "EF Pettersen",
"doi-asserted-by": "publisher",
"first-page": "1605",
"journal-title": "J Comput Chem",
"key": "299_CR40",
"unstructured": "Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1016/j.bbrc.2003.12.081",
"author": "P Prabakaran",
"doi-asserted-by": "publisher",
"first-page": "235",
"issue": "1",
"journal-title": "Biochem Biophys Res Commun",
"key": "299_CR41",
"unstructured": "Prabakaran P, Xiao X, Dimitrov DS (2004) A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem Biophys Res Commun 314(1):235–241. https://doi.org/10.1016/j.bbrc.2003.12.081",
"volume": "314",
"year": "2004"
},
{
"DOI": "10.1016/S1574-1400(05)01009-1",
"doi-asserted-by": "publisher",
"key": "299_CR42",
"unstructured": "Raha K, Merz KMBT-ARC C (2005) Chapter 9 Calculating binding free energy in protein–ligand interaction. In: Annual reports in computational chemistry, vol. 1. Elsevier, pp. 113–130. https://doi.org/10.1016/S1574-1400(05)01009-1"
},
{
"DOI": "10.1128/JVI.02680-07",
"author": "J Shi",
"doi-asserted-by": "publisher",
"first-page": "4620LP",
"issue": "9",
"journal-title": "J Virol",
"key": "299_CR43",
"unstructured": "Shi J, Sivaraman J, Song J (2008) Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J Virol 82(9):4620LP – 4629. https://doi.org/10.1128/JVI.02680-07",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1021/bi702107v",
"author": "J Solowiej",
"doi-asserted-by": "publisher",
"first-page": "2617",
"issue": "8",
"journal-title": "Biochemistry",
"key": "299_CR44",
"unstructured": "Solowiej J, Thomson JA, Ryan K, Luo C, He M, Lou J, Murray BW (2008) Steady-State and pre-steady-state kinetic evaluation of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro Cysteine protease: development of an ion-pair model for catalysis. Biochemistry 47(8):2617–2630. https://doi.org/10.1021/bi702107v",
"volume": "47",
"year": "2008"
},
{
"DOI": "10.1099/0022-1317-69-12-2939",
"author": "W Spaan",
"doi-asserted-by": "publisher",
"first-page": "2939",
"issue": "12",
"journal-title": "J Gen Virol",
"key": "299_CR45",
"unstructured": "Spaan W, Cavanagh D, Horzinek MC (1988) Coronaviruses: structure and genome expression. J Gen Virol 69(12):2939–2952. https://doi.org/10.1099/0022-1317-69-12-2939",
"volume": "69",
"year": "1988"
},
{
"DOI": "10.3390/molecules22071038",
"author": "V Spiwok",
"doi-asserted-by": "publisher",
"journal-title": "Molecules",
"key": "299_CR46",
"unstructured": "Spiwok V (2017) CH/π Interactions in Carbohydrate Recognition. Molecules. https://doi.org/10.3390/molecules22071038",
"year": "2017"
},
{
"DOI": "10.1016/B978-0-12-815422-9.00003-6",
"author": "S Tomar",
"doi-asserted-by": "publisher",
"first-page": "69",
"key": "299_CR47",
"unstructured": "Tomar S, Mudgal R, Pareek A (2019) Chapter 3—RNA-dependent RNA polymerase of alphaviruses: a potential target for the design of drugs against alphaviruses. In: Gupta SPBT-VP (ed) Viral polymerases. Academic Press, pp 69–94. https://doi.org/10.1016/B978-0-12-815422-9.00003-6",
"volume-title": "Viral polymerases",
"year": "2019"
},
{
"DOI": "10.1016/j.antiviral.2020.104762",
"author": "F Touret",
"doi-asserted-by": "publisher",
"first-page": "104762",
"journal-title": "Antivir Res",
"key": "299_CR48",
"unstructured": "Touret F, de Lamballerie X (2020) Of chloroquine and COVID-19. Antivir Res 177:104762. https://doi.org/10.1016/j.antiviral.2020.104762",
"volume": "177",
"year": "2020"
},
{
"DOI": "10.1002/jcc.21334",
"author": "O Trott",
"doi-asserted-by": "publisher",
"first-page": "455",
"issue": "2",
"journal-title": "J Comput Chem",
"key": "299_CR49",
"unstructured": "Trott O, Olson AJ (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1056/NEJMc2004973",
"author": "N van Doremalen",
"doi-asserted-by": "publisher",
"first-page": "1564",
"issue": "16",
"journal-title": "N Engl J Med",
"key": "299_CR50",
"unstructured": "van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ (2020) Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 382(16):1564–1567. https://doi.org/10.1056/NEJMc2004973",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"author": "AC Walls",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Cell",
"key": "299_CR51",
"unstructured": "Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 180:1–12. https://doi.org/10.1016/j.cell.2020.02.058",
"volume": "180",
"year": "2020"
},
{
"DOI": "10.1023/A:1016357811882",
"author": "R Wang",
"doi-asserted-by": "publisher",
"first-page": "11",
"issue": "1",
"journal-title": "J Comput Aided Mol Des",
"key": "299_CR53",
"unstructured": "Wang R, Lai L, Wang S (2002) Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des 16(1):11–26. https://doi.org/10.1023/A:1016357811882",
"volume": "16",
"year": "2002"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"author": "M Wang",
"doi-asserted-by": "publisher",
"first-page": "269",
"issue": "3",
"journal-title": "Cell Res",
"key": "299_CR52",
"unstructured": "Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020a) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30(3):269–271. https://doi.org/10.1038/s41422-020-0282-0",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1002/jmv.25748",
"author": "Y Wang",
"doi-asserted-by": "publisher",
"journal-title": "J Med Virol",
"key": "299_CR54",
"unstructured": "Wang Y, Wang Y, Chen Y, Qin Q (2020b) Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. https://doi.org/10.1002/jmv.25748",
"year": "2020"
},
{
"DOI": "10.1093/nar/gky427",
"author": "A Waterhouse",
"doi-asserted-by": "publisher",
"first-page": "296",
"issue": "1",
"journal-title": "Nucleic Acids Res",
"key": "299_CR55",
"unstructured": "Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(1):296-W303. https://doi.org/10.1093/nar/gky427",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.1093/nar/gkz966",
"doi-asserted-by": "publisher",
"key": "299_CR56",
"unstructured": "WHO (2020) Coronavirus disease (COVID-2019) situation reports 318. https://doi.org/10.1093/nar/gkz966"
},
{
"DOI": "10.1002/pro.3330",
"author": "CJ Williams",
"doi-asserted-by": "publisher",
"first-page": "293",
"issue": "1",
"journal-title": "Protein Sci",
"key": "299_CR57",
"unstructured": "Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB III, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC (2018) MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27(1):293–315. https://doi.org/10.1002/pro.3330",
"volume": "27",
"year": "2018"
},
{
"DOI": "10.1073/pnas.1835675100",
"author": "H Yang",
"doi-asserted-by": "publisher",
"first-page": "13190",
"issue": "23",
"journal-title": "Proc Natl Acad Sci USA",
"key": "299_CR58",
"unstructured": "Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z (2003) The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100(23):13190–13195",
"volume": "100",
"year": "2003"
},
{
"DOI": "10.1093/bioinformatics/btt447",
"author": "J Yang",
"doi-asserted-by": "publisher",
"first-page": "2588",
"issue": "20",
"journal-title": "Bioinformatics",
"key": "299_CR59",
"unstructured": "Yang J, Roy A, Zhang Y (2013) Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29(20):2588–2595. https://doi.org/10.1093/bioinformatics/btt447",
"volume": "29",
"year": "2013"
}
],
"reference-count": 61,
"references-count": 61,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s13721-021-00299-2"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "10"
}