Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin
et al., EMBO Mol. Med., doi:10.15252/emmm.202114122, Nov 2020 (preprint)
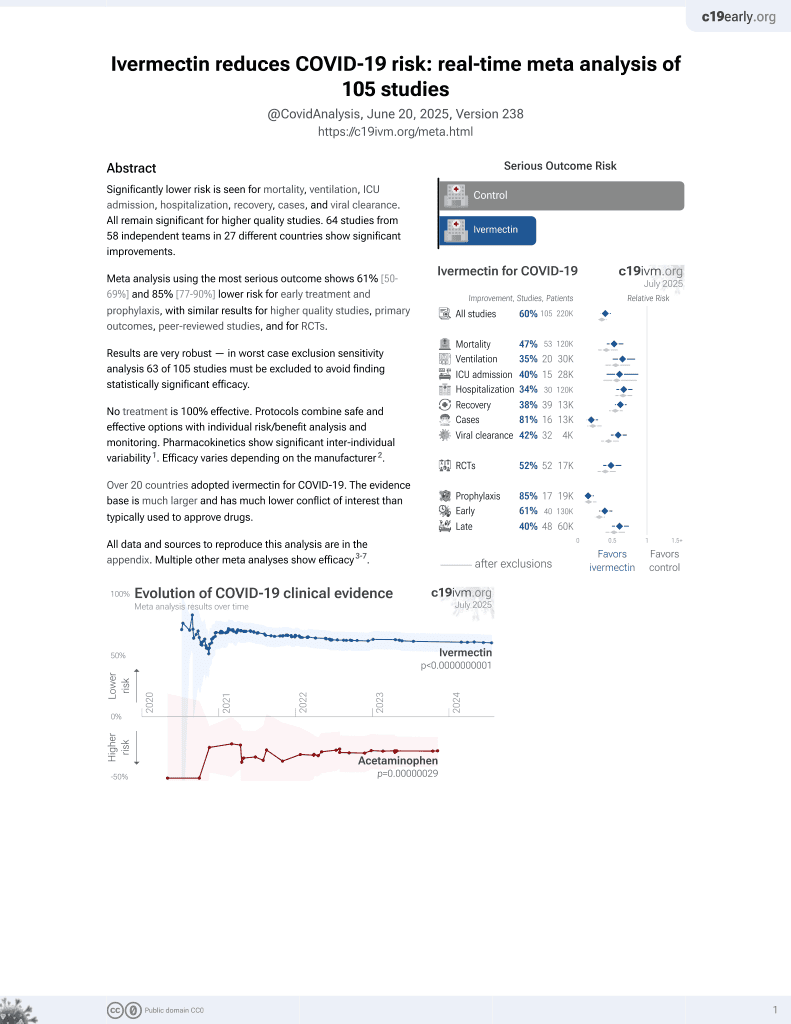
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
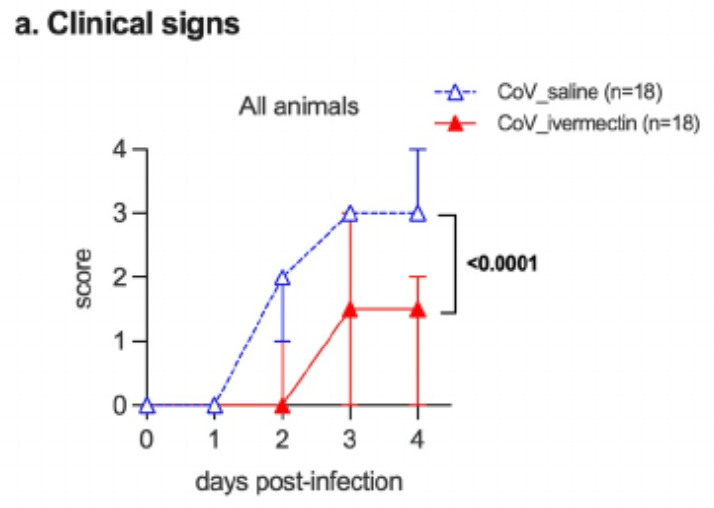
Animal study showing that standard doses of ivermectin prevented clinical deterioration, reduced olfactory deficit, and limited inflammation in the upper and lower respiratory tracts of SARS-CoV-2-infected hamsters.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
de Melo et al., 22 Nov 2020, peer-reviewed, 11 authors.
Attenuation of clinical and immunological outcomes during SARS‐CoV‐2 infection by ivermectin
EMBO Molecular Medicine, doi:10.15252/emmm.202114122
The devastating pandemic due to SARS-CoV-2 and the emergence of antigenic variants that jeopardize the efficacy of current vaccines create an urgent need for a comprehensive understanding of the pathophysiology of COVID-19, including the contribution of inflammation to disease. It also warrants for the search of immunomodulatory drugs that could improve disease outcome. Here, we show that standard doses of ivermectin (IVM), an antiparasitic drug with potential immunomodulatory activities through the cholinergic anti-inflammatory pathway, prevent clinical deterioration, reduce olfactory deficit, and limit the inflammation of the upper and lower respiratory tracts in SARS-CoV-2infected hamsters. Whereas it has no effect on viral load in the airways of infected animals, transcriptomic analyses of infected lungs reveal that IVM dampens type I interferon responses and modulates several other inflammatory pathways. In particular, IVM dramatically reduces the Il-6/Il-10 ratio in lung tissue and promotes macrophage M2 polarization, which might account for the more favorable clinical presentation of IVM-treated animals. Altogether, this study supports the use of immunomodulatory drugs such as IVM, to improve the clinical condition of SARS-CoV-2-infected patients.
Author contributions JPC and HB conceived the experimental hypothesis. GDM, FLaz, FLar, and HB designed the experiments. GDM, FLaz, FLar, LF, LK, SL, AM, and DH performed the experiments. GDM, FLaz, FLar, LF, EK, SL, AM, TC, PP, ML, and P-ML analyzed the data. GDM, J-PC, and HB wrote the manuscript, and all authors edited it.
Conflict of interest The authors declare that they have no conflict of interest.
For more information • COVID-19 section of the WHO website: https://covid19.who.int/
References Aamir K, Khan HU, Sethi G, Hossain MA, Arya A (2020) Wnt signaling mediates TLR pathway and promote unrestrained adipogenesis and
The paper explained
Problem The current pandemic of COVID-19 has caused more than 3.5 million deaths and more than 150 million laboratory-confirmed cases worldwide since December 2019 (as of May 2021). COVID-19, caused by SARS-CoV-2, commonly brings about upper airways and pulmonary symptoms and in severe cases can lead to respiratory distress and death. Different therapeutic approaches have been proposed to fight this disease but comprehensive therapeutic studies are still lacking.
Results We report that ivermectin, used at the standard anti-parasitic dose of 400 µg/kg, protects infected hamsters from developing clinical signs and from losing the sense of smell during SARS-CoV-2 infection. The treated animals exhibited a specific inflammatory response, presenting a reduced type I/III interferon stimulation and a modulation in several..
References
Ar Evalo, Pagotto, Orfido Jl, Daghero, Segovia et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Sci Rep
Bastard, Rosen, Zhang, Michailidis, Hoffmann et al., Autoantibodies against type I IFNs in patients with life-threatening COVID-19, Science
Batalha, Forezi, Lima, Pauli, Boechat et al., Drug repurposing for the treatment of COVID-19: Pharmacological aspects and synthetic approaches, Bioorg Chem
Beco, Petite, Olivry, Comparison of subcutaneous ivermectin and oral moxidectin for the treatment of notoedric acariasis in hamsters, Veterinary Record
Bernigaud, Guillemot, Ahmed-Belkacem, Grimaldi-Bensouda, Lespine et al., Oral ivermectin for a scabies outbreak in a long-term-care facility: potential value in preventing COVID-19 and associated mortality?, Br J Dermatol
Bhaskar, Sinha, Banach, Mittoo, Weissert et al., Cytokine storm in COVID-19-immunopathological mechanisms. Clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper, Front Immunol
Boudewijns, Thibaut, Kaptein, Li, Vergote et al., STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters, Nat Commun
Bray, Rayner, El, Jans, Wagstaff, Ivermectin and COVID-19: a report in antiviral research, widespread interest, an FDA warning, two letters to the editor and the authors' responses, Antiviral Res
Calabrese, Kozumbo, Kapoor, Dhawan, Jimenez et al., NRF2 activation putatively mediates clinical benefits of low-dose radiotherapy in COVID-19 pneumonia and acute respiratory distress syndrome (ards): novel mechanistic considerations, Radiother Oncol
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res
Camprubí, Almuedo-Riera, Mart I-Soler, Soriano, Hurtado et al., Lack of efficacy of standard doses of ivermectin in severe COVID-19 patients, PLoS One
Cerdan, Sisquellas, Pereira, Gomes, Changeux et al., The glycine receptor allosteric ligands library (GRALL), Bioinformatics
Cereda, Tirani, Rovida, Demicheli, Ajelli et al., The early phase of the COVID-19 outbreak in Lombardy
Chaccour, Casellas, Blanco-Di Matteo, Pineda, Fernandez-Montero et al., The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: a pilot, double-blind, placebo-controlled, randomized clinical trial, EClinicalMedicine
Chaccour, Hammann, On-Garc Ia, Rabinovich, Ivermectin and COVID-19: keeping rigor in times of urgency, Am J Trop Med Hyg
Chan, Zhang, Yuan, Poon, Chan et al., Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility, Clin Infect Dis
Changeux, Amoura, Rey, Miyara, A nicotinic hypothesis for Covid-19 with preventive and therapeutic implications, CR Biol
Chen, Pan, Anatomical and pathological observation and analysis of SARS and COVID-19: microthrombosis is the main cause of death, Biol Proced Online
Chen, Tan, Kou, Duan, Wang et al., Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool, BMC Bioinformatics
Cokelaer, Desvillechabrol, Legendre, Cardon, Sequana': a set of snakemake NGS pipelines, J Open Source Softw
Cokelaer, Pultz, Harder, Serra-Musach, Saez-Rodriguez, BioServices: a common Python package to access biological Web Services programmatically, Bioinformatics
Cross, Linker, Leslie, Sex-dependent effects of nicotine on the developing brain, J Neurosci Res
De Melo, Lazarini, Levallois, Hautefort, Michel et al., COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters, Sci Transl Med
Dey, Sen, Maulik, Unveiling COVID-19-associated organ-specific cell types and cell-specific pathway cascade, Brief Bioinform
Dobin, Davis, Schlesinger, Drenkow, Zaleski et al., STAR: ultrafast universal RNA-seq aligner, Bioinformatics
Ewels, Magnusson, Lundin, MultiQC: summarize analysis results for multiple tools and samples in a single report, Bioinformatics
Gahring, Myers, Dunn, Weiss, Rogers, Nicotinic alpha 7 receptor expression and modulation of the lung epithelial response to lipopolysaccharide, PLoS One
Galani, Triantafyllia, Eleminiadou, Koltsida, Stavropoulos et al., Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness, Immunity
Gianchandani, Esfandiari, Ang, Iyengar, Knotts et al., Managing hyperglycemia in the COVID-19 inflammatory storm, Diabetes
Guzzo, Furtek, Porras, Chen, Tipping et al., Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects, J Clin Pharmacol
Han, Mukdad, Long, Lopez, Anosmia in COVID-19: mechanisms and significance, Chem Senses
Hanafi, Szumlas, Fryauff, El-Hossary, Singer et al., Effects of ivermectin on bloodfeeding Phlebotomus papatasi, and the promastigote stage of Leishmania major, Vector Borne Zoonotic Dis
Hasanoglu, Korukluoglu, Asilturk, Cosgun, Kalem et al., Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg, Infection
Heidary, Gharebaghi, Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen, J Antibiot
Hibbs, Gouaux, Principles of activation and permeation in an anion-selective Cys-loop receptor, Nature
Hill, Abdulamir, Ahmed, Asghar, Babalola et al., Meta-analysis of randomized trials of ivermectin to treat SARS-CoV-2 infection, Res Squ, doi:10.21203/rs.3.rs-148845/v1
Hoagland, Møller, Uhl, Oishi, Frere et al., Leveraging the antiviral type-I interferon system as a first line defense against SARS-CoV-2 pathogenicity, Immunity
Horby, Lim, Emberson, Mafham, Bell et al., Dexamethasone in hospitalized patients with Covid-19 -preliminary report, N Engl J Med
Hu, Cai, Song, Li, Zhao et al., Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor, Future Virol
Huntley, Sawford, Mutowo-Meullenet, Shypitsyna, Bonilla et al., The GOA database: gene ontology annotation updates for 2015, Nucleic Acids Res
Isidori, Giannetta, Pofi, Venneri, Gianfrilli et al., Targeting the NO-cGMP-PDE5 pathway in COVID-19 infection. The DEDALO project, Andrology
Islam, Rahman, Aydin, Beklen, Arga et al., Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19, Eur J Pharmacol
Jermain, Hanafin, Cao, Lifschitz, Lanusse et al., Development of a minimal physiologically-based pharmacokinetic model to simulate lung exposure in humans following oral administration of ivermectin for COVID-19 drug repurposing, J Pharm Sci
Jin, Bai, He, Wu, Liu et al., Gender differences in patients with COVID-19: focus on severity and mortality, Frontiers in Public Health
Kanehisa, Goto, KEGG: kyoto encyclopedia of genes and genomes, Nucleic Acids Res
Kaur, Shekhar, Sharma, Sarma, Prakash et al., Ivermectin as a potential drug for treatment of COVID-19: an in-sync review with clinical and computational attributes, Pharmacol Rep
Klinkhammer, Schnepf, Ye, Schwaderlapp, Gad et al., IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission, eLife
Kory, Meduri, Varon, Iglesias, Marik, Review of the emerging evidence demonstrating the efficacy of ivermectin in the prophylaxis and treatment of COVID-19, Am J Ther
Krause, Buisson, Bertrand, Corringer, Galzi et al., Ivermectin: a positive allosteric effector of the alpha7 neuronal nicotinic acetylcholine receptor, Mol Pharmacol
Köster, Rahmann, Snakemake-a scalable bioinformatics workflow engine, Bioinformatics
Laing, Devaney, Ivermectin -old drug, new tricks?, Trends Parasitol
Lazarini, Gabellec, Torquet, Lledo, Early activation of microglia triggers long-lasting impairment of adult neurogenesis in the olfactory bulb, The Journal of Neuroscience
Li, Zhao, Zhan, Quantitative proteomics reveals a broadspectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J Cell Physiol
Liao, Smyth, Shi, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics
Lifschitz, Virkel, Sallovitz, Sutra, Galtier et al., Comparative distribution of ivermectin and doramectin to parasite location tissues in cattle, Vet Parasitol
Love, Huber, Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol
Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet J
Masood, Mahmood, Shahid, Nasir, Ghanchi et al., Transcriptomic profiling of disease severity in patients with COVID-19 reveals role of blood clotting and vasculature related genes, doi:10.1101/2020.06.18.20132571
Mcelvaney, Hobbs, Qiao, Mcelvaney, Moll et al., A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19, EBioMedicine
Mcelvaney, Mcevoy, Mcelvaney, Carroll, Murphy et al., Characterization of the inflammatory response to severe COVID-19 illness, Am J Respir Crit Care Med
Melotti, Mas, Kuciak, Lorente-Trigos, Borges et al., The river blindness drug Ivermectin and related macrocyclic lactones inhibit WNT-TCF pathway responses in human cancer, EMBO Mol Med
Mi, Muruganujan, Ebert, Huang, Thomas, PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools, Nucleic Acids Res
Muñoz-Fontela, Dowling, Funnell, Gsell, Ax et al., Animal models for COVID-19, Nature
Oczypok, Perkins, Oury, All the "RAGE" in lung disease: The receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses, Paediatr Respir Rev
Okabayashi, Kojima, Masaki, Yokota, Imaizumi et al., Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells, Virus Res
Oliviero, De Castro, Coperchini, Chiovato, Rotondi, COVID-19 pulmonary and olfactory dysfunctions: is the chemokine CXCL10 the common denominator?, The Neuroscientist
Pavlov, Tracey, The vagus nerve and the inflammatory reflexlinking immunity and metabolism, The Authors EMBO Molecular Medicine
Pei, Xiao, Guo, Pei, Wei et al., Sustained stimulation of β(2)AR inhibits insulin signaling in H9C2 cardiomyoblast cells through the PKA-dependent signaling pathway, Diabetes Metab Syndr Obes
Pfaffl, A new mathematical model for relative quantification in real-time RT-PCR, Nucleic Acids Res
Potus, Mai, Lebret, Malenfant, Breton-Gagnon et al., Novel insights on the pulmonary vascular consequences of COVID-19, Am J Physiol Lung Cell Mol Physiol
Qiu, Cui, Hautefort, Haehner, Zhao et al., Olfactory and gustatory dysfunction as an early identifier of COVID-19 in adults and children: an international multicenter study, Otolaryngol Head Neck Surg
Rajter, Sherman, Fatteh, Vogel, Sacks et al., Use of ivermectin is associated with lower mortality in hospitalized patients with COVID-19 (ICON study), Chest
Raker, Becker, Steinbrink, The cAMP pathway as therapeutic target in autoimmune and inflammatory diseases, Front Immunol
Raslan, Yoon, WNT signaling in lung repair and regeneration, Mol Cells
Rossotti, Travi, Ughi, Corradin, Baiguera et al., Safety and efficacy of anti-il6-receptor tocilizumab use in severe and critical patients affected by coronavirus disease 2019: A comparative analysis, J Infect
Said, Glutamate receptors and asthmatic airway disease, Trends Pharmacol Sci
Sajid, Iqbal, Muhammad, Iqbal, Immunomodulatory effect of various anti-parasitics: a review, Parasitology
Samuel, Majd, Richter, Ghazizadeh, Zekavat et al., Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men, Cell Stem Cell
Sang, Miller, Blecha, Macrophage polarization in virus-host interactions, J Clin Cell Immunol
Scully, Haverfield, Ursin, Tannenbaum, Klein, Considering how biological sex impacts immune responses and COVID-19 outcomes, Nat Rev Immunol
Sia, Yan, Chin, Fung, Choy et al., Pathogenesis and transmission of SARS-CoV-2 in golden hamsters, Nature
Stanifer, Guo, Doldan, Boulant, Importance of type I and III interferons at respiratory and intestinal barrier surfaces, Front Immunol
Sun, Zhuang, Zheng, Li, Wong et al., Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment, Cell
Suo, Liu, Feng, Guo, Hu et al., ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens, Emerg Microbes Infect
Takahashi, Ellingson, Wong, Israelow, Lucas et al., Sex differences in immune responses that underlie COVID-19 disease outcomes, Nature
Tizabi, Getachew, Copeland, Aschner, Nicotine and the nicotinic cholinergic system in COVID-19, FEBS J
Vaduganathan, Vardeny, Michel, Mcmurray, Pfeffer et al., Renin-angiotensin-aldosterone system inhibitors in patients with covid-19, N Engl J Med
Van Den Eynden, Sahebali, Horwood, Carmans, Brône et al., Glycine and glycine receptor signalling in non-neuronal cells, Front Mol Neurosci
Varet, Brillet-Gu Eguen, Copp Ee, Dillies, SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data
Virgiliis, Giovanni, Lung innervation in the eye of a cytokine storm: neuroimmune interactions and COVID-19, Nat Rev Neurol
Vom Steeg, Klein, Sex and sex steroids impact influenza pathogenesis across the life course, Semin Immunopathol
Xydakis, Dehgani-Mobaraki, Holbrook, Geisthoff, Bauer et al., Smell and taste dysfunction in patients with COVID-19, Lancet Infect Dis
Zemkova, Tvrdonova, Bhattacharya, Jindrichova, Allosteric modulation of ligand gated ion channels by ivermectin, Physiol Res
Zhang, Bastard, Liu, Pen, Velez et al., Inborn errors of type I IFN immunity in patients with life-threatening COVID-19, Science
Zhang, Liu, Wang, Yang, Li et al., SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19, J Hematol Oncol
Zhao, Yu, Lv, Meng, Xu et al., Activation of alpha-7 nicotinic acetylcholine receptors (α7nAchR) promotes the protective autophagy in LPS-induced acute lung injury (ALI) in vitro and in vivo, Inflammation
Zhe, Hongyuan, Wenjuan, Peng, Xiaowei et al., Blockade of glutamate receptor ameliorates lipopolysaccharide-induced sepsis through regulation of neuropeptides, Biosci Rep
DOI record:
{
"DOI": "10.15252/emmm.202114122",
"ISSN": [
"1757-4676",
"1757-4684"
],
"URL": "http://dx.doi.org/10.15252/emmm.202114122",
"alternative-id": [
"10.15252/emmm.202114122"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2021-02-11"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2021-06-23"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2021-07-12"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-0747-7760",
"affiliation": [
{
"name": "Lyssavirus Epidemiology and Neuropathology Unit Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "de Melo",
"given": "Guilherme Dias",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-5572-6982",
"affiliation": [
{
"name": "Perception and Memory Unit Institut Pasteur CNRS UMR 3571 Paris France"
}
],
"authenticated-orcid": false,
"family": "Lazarini",
"given": "Françoise",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0881-4263",
"affiliation": [
{
"name": "Lyssavirus Epidemiology and Neuropathology Unit Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "Larrous",
"given": "Florence",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Lyssavirus Epidemiology and Neuropathology Unit Institut Pasteur Paris France"
}
],
"family": "Feige",
"given": "Lena",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Biomics Technological Platform Center for Technological Resources and Research (C2RT) Institut Pasteur Paris France"
},
{
"name": "Bioinformatics and Biostatistics Hub Computational Biology Department Institut Pasteur Paris France"
}
],
"family": "Kornobis",
"given": "Etienne",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5805-1092",
"affiliation": [
{
"name": "Biology of Infection Unit Institut Pasteur Inserm U1117 Paris France"
}
],
"authenticated-orcid": false,
"family": "Levallois",
"given": "Sylvain",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Nuclear Organization and Oncogenesis Unit Institut Pasteur Paris France"
}
],
"family": "Marchio",
"given": "Agnès",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5609-4398",
"affiliation": [
{
"name": "Lyssavirus Epidemiology and Neuropathology Unit Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "Kergoat",
"given": "Lauriane",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5874-4377",
"affiliation": [
{
"name": "Experimental Neuropathology Unit Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "Hardy",
"given": "David",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6286-1138",
"affiliation": [
{
"name": "Biomics Technological Platform Center for Technological Resources and Research (C2RT) Institut Pasteur Paris France"
},
{
"name": "Bioinformatics and Biostatistics Hub Computational Biology Department Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "Cokelaer",
"given": "Thomas",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Nuclear Organization and Oncogenesis Unit Institut Pasteur Paris France"
}
],
"family": "Pineau",
"given": "Pascal",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4491-1063",
"affiliation": [
{
"name": "Biology of Infection Unit Institut Pasteur Inserm U1117 Paris France"
},
{
"name": "Division of Infectious Diseases and Tropical Medicine Institut Imagine Université de Paris Necker‐Enfants Malades University Hospital AP‐HP Paris France"
}
],
"authenticated-orcid": false,
"family": "Lecuit",
"given": "Marc",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Perception and Memory Unit Institut Pasteur CNRS UMR 3571 Paris France"
}
],
"family": "Lledo",
"given": "Pierre‐Marie",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0297-1583",
"affiliation": [
{
"name": "Neuroscience Department Institut Pasteur Collège de France Paris France"
}
],
"authenticated-orcid": false,
"family": "Changeux",
"given": "Jean‐Pierre",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2608-5589",
"affiliation": [
{
"name": "Lyssavirus Epidemiology and Neuropathology Unit Institut Pasteur Paris France"
}
],
"authenticated-orcid": false,
"family": "Bourhy",
"given": "Hervé",
"sequence": "additional"
}
],
"container-title": "EMBO Molecular Medicine",
"container-title-short": "EMBO Mol Med",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"www.embopress.org"
]
},
"created": {
"date-parts": [
[
2021,
6,
25
]
],
"date-time": "2021-06-25T12:02:32Z",
"timestamp": 1624622552000
},
"deposited": {
"date-parts": [
[
2023,
12,
18
]
],
"date-time": "2023-12-18T20:56:16Z",
"timestamp": 1702932976000
},
"funder": [
{
"DOI": "10.13039/501100003762",
"doi-asserted-by": "publisher",
"name": "Institut Pasteur"
}
],
"indexed": {
"date-parts": [
[
2024,
5,
12
]
],
"date-time": "2024-05-12T01:15:22Z",
"timestamp": 1715476522203
},
"is-referenced-by-count": 37,
"issue": "8",
"issued": {
"date-parts": [
[
2021,
7,
12
]
]
},
"journal-issue": {
"issue": "8",
"published-print": {
"date-parts": [
[
2021,
8,
9
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
7,
12
]
],
"date-time": "2021-07-12T00:00:00Z",
"timestamp": 1626048000000
}
}
],
"link": [
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.15252/emmm.202114122",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/full-xml/10.15252/emmm.202114122",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.embopress.org/doi/pdf/10.15252/emmm.202114122",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2021,
7,
12
]
]
},
"published-online": {
"date-parts": [
[
2021,
7,
12
]
]
},
"published-print": {
"date-parts": [
[
2021,
8,
9
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/j.phrs.2019.104602",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_2_1"
},
{
"DOI": "10.1038/s41598-021-86679-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_3_1"
},
{
"author": "AVMA",
"key": "e_1_2_11_4_1",
"volume-title": "AVMA guidelines for the euthanasia of animals: 2020 edition*",
"year": "2020"
},
{
"article-title": "Viral concentration determination through plaque assays: using traditional and novel overlay systems",
"author": "Baer A",
"first-page": "e52065",
"journal-title": "J Vis Exp",
"key": "e_1_2_11_5_1",
"year": "2014"
},
{
"DOI": "10.1126/science.abd4585",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_6_1"
},
{
"DOI": "10.1016/j.bioorg.2020.104488",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_7_1"
},
{
"DOI": "10.1136/vr.149.11.324",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_8_1"
},
{
"DOI": "10.1111/bjd.19821",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_9_1"
},
{
"DOI": "10.3389/fimmu.2020.01648",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_10_1"
},
{
"DOI": "10.1038/s41467-020-19684-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_11_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104805",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_12_1"
},
{
"DOI": "10.1016/j.radonc.2021.04.015",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_13_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_14_1"
},
{
"DOI": "10.1371/journal.pone.0242184",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_15_1"
},
{
"DOI": "10.1093/bioinformatics/btaa170",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_16_1"
},
{
"article-title": "The early phase of the COVID‐19 outbreak in Lombardy, Italy",
"author": "Cereda D",
"journal-title": "arXiv",
"key": "e_1_2_11_17_1",
"year": "2020"
},
{
"DOI": "10.4269/ajtmh.20-0271",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_18_1"
},
{
"DOI": "10.1016/j.eclinm.2020.100720",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_19_1"
},
{
"article-title": "Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID‐19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility",
"author": "Chan JF",
"first-page": "2428",
"journal-title": "Clin Infect Dis",
"key": "e_1_2_11_20_1",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.5802/crbiol.8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_21_1"
},
{
"DOI": "10.1186/1471-2105-14-128",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_22_1"
},
{
"DOI": "10.1186/s12575-021-00142-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_23_1"
},
{
"DOI": "10.1093/bioinformatics/btt547",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_24_1"
},
{
"DOI": "10.21105/joss.00352",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_25_1"
},
{
"DOI": "10.1002/jnr.23878",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_26_1"
},
{
"DOI": "10.1038/s41582-020-0402-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_27_1"
},
{
"DOI": "10.1093/bib/bbaa214",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_28_1"
},
{
"DOI": "10.1093/bioinformatics/bts635",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_29_1"
},
{
"DOI": "10.1093/bioinformatics/btw354",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_30_1"
},
{
"DOI": "10.1371/journal.pone.0175367",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_31_1"
},
{
"DOI": "10.1016/j.immuni.2017.04.025",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_32_1"
},
{
"DOI": "10.2337/dbi20-0022",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_33_1"
},
{
"DOI": "10.1177/009127002237994",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_34_1"
},
{
"DOI": "10.1093/chemse/bjaa040",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_35_1"
},
{
"DOI": "10.1089/vbz.2009.0030",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_36_1"
},
{
"DOI": "10.1007/s15010-020-01548-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_37_1"
},
{
"DOI": "10.1038/s41429-020-0336-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_38_1"
},
{
"DOI": "10.1038/nature10139",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_39_1"
},
{
"article-title": "Meta‐analysis of randomized trials of ivermectin to treat SARS‐CoV‐2 infection",
"author": "Hill A",
"journal-title": "Res Squ",
"key": "e_1_2_11_40_1",
"year": "2021"
},
{
"DOI": "10.1016/j.immuni.2021.01.017",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_41_1"
},
{
"article-title": "Dexamethasone in hospitalized patients with Covid‐19 ‐ preliminary report",
"author": "Horby P",
"first-page": "693",
"journal-title": "N Engl J Med",
"key": "e_1_2_11_42_1",
"volume": "384",
"year": "2020"
},
{
"DOI": "10.2217/fvl-2020-0099",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_43_1"
},
{
"DOI": "10.1093/nar/gku1113",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_44_1"
},
{
"DOI": "10.1111/andr.12837",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_45_1"
},
{
"DOI": "10.1016/j.ejphar.2020.173594",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_46_1"
},
{
"DOI": "10.1016/j.xphs.2020.08.024",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_47_1"
},
{
"DOI": "10.3389/fpubh.2020.00152",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_48_1"
},
{
"DOI": "10.1093/nar/28.1.27",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_49_1"
},
{
"DOI": "10.1007/s43440-020-00195-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_50_1"
},
{
"DOI": "10.7554/eLife.33354",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_51_1"
},
{
"DOI": "10.1097/MJT.0000000000001377",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_52_1"
},
{
"DOI": "10.1093/bioinformatics/bts480",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_53_1"
},
{
"DOI": "10.1124/mol.53.2.283",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_54_1"
},
{
"DOI": "10.1016/j.pt.2017.02.004",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_55_1"
},
{
"DOI": "10.1523/JNEUROSCI.6394-11.2012",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_56_1"
},
{
"DOI": "10.1002/jcp.30055",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_57_1"
},
{
"DOI": "10.1093/bioinformatics/btt656",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_58_1"
},
{
"DOI": "10.1016/S0304-4017(99)00175-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_59_1"
},
{
"DOI": "10.1186/s13059-014-0550-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_60_1"
},
{
"DOI": "10.14806/ej.17.1.200",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_61_1"
},
{
"article-title": "Transcriptomic profiling of disease severity in patients with COVID‐19 reveals role of blood clotting and vasculature related genes",
"author": "Masood KI",
"journal-title": "medRxiv",
"key": "e_1_2_11_62_1",
"year": "2020"
},
{
"DOI": "10.1016/j.ebiom.2020.103026",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_63_1"
},
{
"DOI": "10.1164/rccm.202005-1583OC",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_64_1"
},
{
"DOI": "10.1126/scitranslmed.abf8396",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_65_1"
},
{
"DOI": "10.15252/emmm.201404084",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_66_1"
},
{
"DOI": "10.1093/nar/gky1038",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_67_1"
},
{
"DOI": "10.1038/s41586-020-2787-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_68_1"
},
{
"DOI": "10.1016/j.prrv.2017.03.012",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_69_1"
},
{
"DOI": "10.1016/j.virusres.2011.07.011",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_70_1"
},
{
"DOI": "10.1177/1073858420939033",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_71_1"
},
{
"DOI": "10.1038/nrendo.2012.189",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_72_1"
},
{
"DOI": "10.2147/DMSO.S268028",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_73_1"
},
{
"DOI": "10.1093/nar/29.9.e45",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_74_1"
},
{
"DOI": "10.1152/ajplung.00195.2020",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_75_1"
},
{
"DOI": "10.1177/0194599820934376",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_76_1"
},
{
"DOI": "10.1016/j.chest.2020.10.009",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_77_1"
},
{
"DOI": "10.3389/fimmu.2016.00123",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_78_1"
},
{
"article-title": "WNT signaling in lung repair and regeneration",
"author": "Raslan AA",
"first-page": "774",
"journal-title": "Mol Cells",
"key": "e_1_2_11_79_1",
"volume": "43",
"year": "2020"
},
{
"DOI": "10.1016/j.jinf.2020.07.008",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_80_1"
},
{
"DOI": "10.1016/S0165-6147(98)01275-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_81_1"
},
{
"DOI": "10.1017/S0031182005009108",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_82_1"
},
{
"DOI": "10.1016/j.stem.2020.11.009",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_83_1"
},
{
"article-title": "Macrophage polarization in virus‐host interactions",
"author": "Sang Y",
"first-page": "311",
"journal-title": "J Clin Cell Immunol",
"key": "e_1_2_11_84_1",
"volume": "6",
"year": "2015"
},
{
"DOI": "10.1038/s41577-020-0348-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_85_1"
},
{
"DOI": "10.1038/s41586-020-2342-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_86_1"
},
{
"DOI": "10.3389/fimmu.2020.608645",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_87_1"
},
{
"DOI": "10.1007/s00281-018-0718-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_88_1"
},
{
"DOI": "10.1016/j.cell.2020.06.010",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_89_1"
},
{
"DOI": "10.1080/22221751.2020.1772678",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_90_1"
},
{
"DOI": "10.1038/s41586-020-2700-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_91_1"
},
{
"DOI": "10.1111/febs.15521",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_92_1"
},
{
"DOI": "10.1056/NEJMsr2005760",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_93_1"
},
{
"DOI": "10.3389/neuro.02.009.2009",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_94_1"
},
{
"DOI": "10.1371/journal.pone.0157022",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_95_1"
},
{
"author": "WHO",
"key": "e_1_2_11_96_1",
"volume-title": "Protocol: real‐time RT‐PCR assays for the detection of SARS‐CoV‐2",
"year": "2020"
},
{
"DOI": "10.1016/S1473-3099(20)30293-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_97_1"
},
{
"DOI": "10.33549/physiolres.932711",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_98_1"
},
{
"DOI": "10.1126/science.abd4570",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_99_1"
},
{
"DOI": "10.1186/s13045-020-00954-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_100_1"
},
{
"DOI": "10.1007/s10753-019-01088-w",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_101_1"
},
{
"DOI": "10.1042/BSR20171629",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_102_1"
}
],
"reference-count": 101,
"references-count": 101,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2020.11.21.392639",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://www.embopress.org/doi/10.15252/emmm.202114122"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Attenuation of clinical and immunological outcomes during SARS‐CoV‐2 infection by ivermectin",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy",
"volume": "13"
}