Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases
et al., ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1, Apr 2024
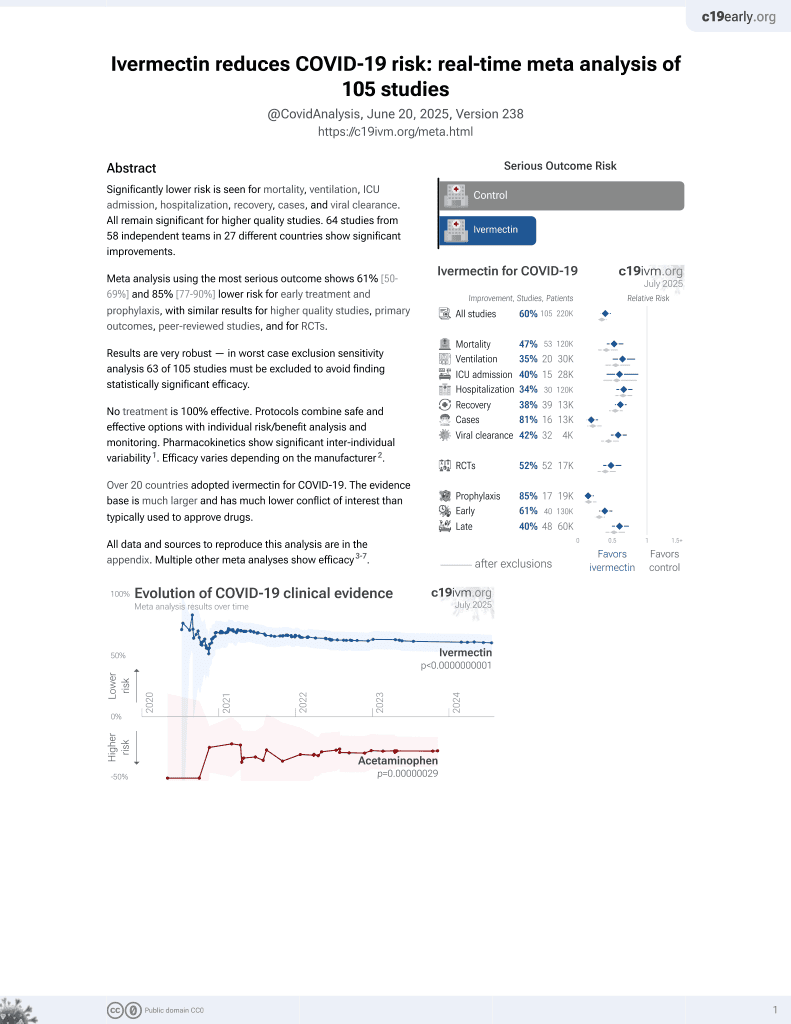
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico study identifying potential drugs beneficial for COVID-19 by integrating transcriptomics, proteomics, metabolomics, lipidomics, and drug data. Authors explore interactions between drugs, molecular features, and disease severity. Hypothesis-driven analysis, using IL-6 and IL-6R as seeds, highlighted immunosuppressants, corticosteroids, and IL-6 inhibitors as promising candidates, while data-driven analysis, using STAT1, SOD2, and lipid/metabolite markers as seeds, identified antioxidants, kinase inhibitors, and protein synthesis inhibitors. Network analysis revealed key hubs like CCL2, CCL4, NFKB1, and HGF that interact with drug candidates and influence disease progression. Treatments identified in the top 20 lists include ivermectin, zinc, azithromycin, indomethacin, curcumin, vitamin C, metformin, mebendazole, and acetylcysteine.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
Study covers ivermectin, zinc, indomethacin, curcumin, vitamin C, metformin, N-acetylcysteine, and mebendazole.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Agamah et al., 16 Apr 2024, preprint, 6 authors.
Contact: francisagamahh@gmail.com, peter-bram.thoen@radboudumc.nl, emile.chimusa@northumbria.ac.uk.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for
Background: The development and roll-out of vaccines, and the use of various drugs have contributed to controlling the COVID-19 pandemic. Nevertheless, challenges such as the inequitable distribution of vaccines, the influence of emerging viral lineages and immune evasive variants on vaccine efficacy, and the inadequate immune defense in subgroups of the population continue to motivate the development of new drugs to combat the disease. Aim: In this study, we sought to identify, prioritize, and characterize drug repurposing candidates appropriate for treating mild, moderate, or severe COVID-19 using a network-based integrative approach that systematically integrates drug-related data and multi-omics datasets.
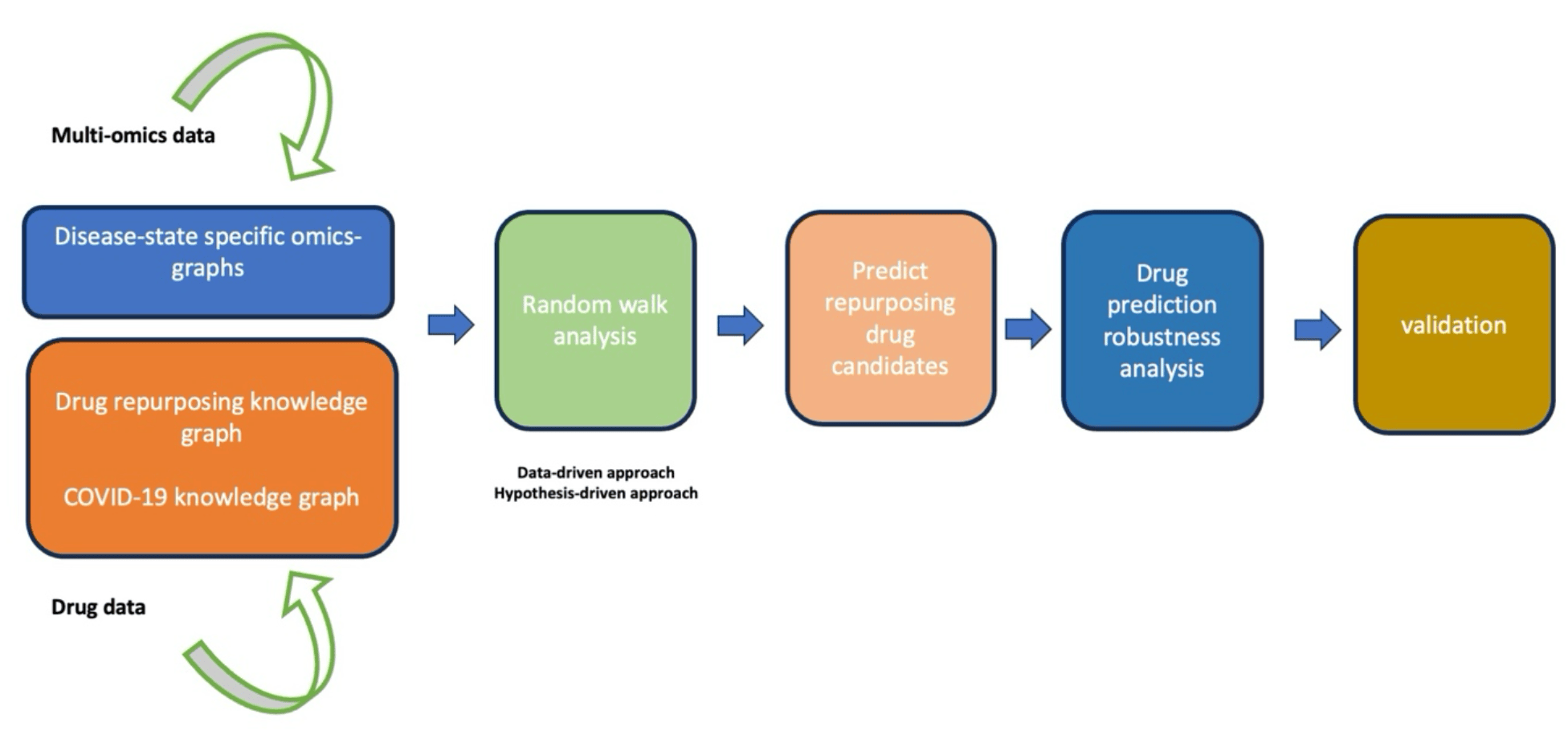
Methods: We leveraged drug data, and multi-omics data, and used a random walk restart algorithm to explore an integrated knowledge graph comprised of three subgraphs: (i) a COVID-19 knowledge graph, (ii) a drug repurposing knowledge graph, and (iii) a COVID-19 disease-state specific omics graph.
Results: We prioritized twenty FDA-approved agents as potential candidate drugs for mild, moderate, and severe COVID-19 disease phases. Specifically, drugs that could stimulate immune cell recruitment and activation including histamine, curcumin, and paclitaxel have potential utility in mild disease states to mitigate disease progression. Drugs like omacetaxine, crizotinib, and vorinostat that exhibit antiviral properties and have the potential to inhibit viral replication can be considered for mild to moderate COVID-19 disease states. Also, given the association between antioxidant deficiency and high inflammatory factors that trigger cytokine storms, antioxidants like glutathione can be considered for moderate disease states. Drugs that exhibit potent antiinflammatory effects like (i) anti-inflammatory drugs (sarilumab and tocilizumab), (ii) corticosteroids (dexamethasone and hydrocortisone), and (iii) immunosuppressives (sirolimus and cyclosporine) are potential candidates for moderate to severe disease states that trigger a hyperinflammatory cascade of COVID-19.
Conclusion: Our study demonstrates that the multi-omics data-driven integrative analysis within the drug data enables prioritizing drug candidates for COVID-19 disease phases, offering a comprehensive basis for therapeutic strategies that can be brought to market quickly given their established safety profiles. Importantly, the multi-omics data-driven integrative analysis within the drug data approach implemented here can be used to prioritize drug repurposing candidates appropriate for other diseases.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication The authors have consented for the work to be published.
Supplementary Files
Supplementary data Supplementary file 1 Supplementary file 2 Supplementary file 3 Supplementary file 4 Supplementary file 5 Next, we prioritized and characterized candidate drugs followed by drug prediction robustness analysis. Finally, we concluded the analysis by validating the predicted drug candidates. Figure legends
References
Agamah, Computational approaches for network-based integrative multiomics analysis, Frontiers in Molecular Biosciences
Agamah, Network-based integrative multi-omics approach reveals biosignatures specific to COVID-19 disease phases, bioRxiv
Al-Kuraishy, Anti-histamines and Covid-19: hype or hope, JPMA J Pak Med Assoc
Alam, Czajkowsky, SARS-CoV-2 infection and oxidative stress: Pathophysiological insight into thrombosis and therapeutic opportunities, Cytokine & Growth Factor Reviews
Aoyagi, Case Report: Successful Treatment of Five Critically Ill Coronavirus Disease 2019 Patients Using Combination Therapy With Etoposide and Corticosteroids, Frontiers in Medicine
Arazi, Human systems immunology: hypothesis-based modeling and unbiased data-driven approaches
Archin, Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy, Nature
Attademo, Bernardini, Are dopamine and serotonin involved in COVID-19 pathophysiology?, The European journal of psychiatry
Bailly, Etoposide: A rider on the cytokine storm, Cytokine
Baptista, Brière, Baudot, Random Walk with Restart on multilayer networks: from node prioritisation to supervised link prediction and beyond, BMC bioinformatics
Baptista, Gonzalez, Baudot, Universal multilayer network exploration by random walk with restart, Communications Physics
Battaglia, Rapamycin and interleukin-10 treatment induces T regulatory type 1 cells that mediate antigen-specific transplantation tolerance, Diabetes
Beigel, Remdesivir for the treatment of Covid-19, New England Journal of Medicine
Belli, Protective role of tacrolimus, deleterious role of age and comorbidities in liver transplant recipients with Covid-19: results from the ELITA/ELTR multi-center European study, Gastroenterology
Ben Abdallah, Twice-daily oral zinc in the treatment of patients with coronavirus disease 2019: a randomized double-blind controlled trial, Clinical Infectious Diseases
Bengtson, An open label trial to assess safety of losartan for treating worsening respiratory illness in COVID-19, Frontiers in medicine
Benucci, COVID-19 pneumonia treated with Sarilumab: A clinical series of eight patients
Benucci, COVID-19 pneumonia treated with sarilumab: a clinical series of eight patients, Journal of medical virology
Bharadwaj, SARS-CoV-2 Mpro inhibitors: identification of anti-SARS-CoV-2 Mpro compounds from FDA approved drugs, Journal of Biomolecular Structure and Dynamics
Bhimraj, Infectious Diseases Society of America Guidelines on the treatment and management of patients with coronavirus disease 2019 (COVID-19), Clinical Infectious Diseases
Boor, Rapamycin has suppressive and stimulatory effects on human plasmacytoid dendritic cell functions, Clinical & Experimental Immunology
Bramante, Outpatient treatment of COVID-19 and incidence of post-COVID-19 condition over 10 months (COVID-OUT): a multicentre, randomised, quadruple-blind, parallel-group, phase 3 trial, The Lancet Infectious Diseases
Bramante, Outpatient treatment of COVID-19 and incidence of post-COVID-19 condition over 10 months (COVID-OUT): a multicentre, randomised, quadruple-blind, parallel-group, phase 3 trial, The Lancet Infectious Diseases
Bustamante, Tryptophan Metabolism 'Hub'Gene Expression Associates with Increased Inflammation and Severe Disease Outcomes in COVID-19 Infection and Inflammatory Bowel Disease, International Journal of Molecular Sciences
Carlos, Histamine modulates mast cell degranulation through an indirect mechanism in a model IgE-mediated reaction, European journal of immunology
Cecchini, Cecchini ; Pono, COVID-19 infection and oxidative stress: an under-explored approach for prevention and treatment? The Pan African Medical Journal, Archives of medical research
Cengiz, Effect of oral l-Glutamine supplementation on Covid-19 treatment, Clinical nutrition experimental
Chen, Confronting the controversy: interleukin-6 and the COVID-19 cytokine storm syndrome, Eur Respiratory Soc
Chen, Immunomodulatory and antiviral activity of metformin and its potential implications in treating coronavirus disease 2019 and lung injury, Frontiers in Immunology
Chow, Association of early aspirin use with in-hospital mortality in patients with moderate COVID-19, JAMA network open
Chowdhury, Pathak, Neuroprotective immunity by essential nutrient "Choline" for the prevention of SARS CoV2 infections: An in silico study by molecular dynamics approach, Chemical physics letters
Consortium, Repurposed antiviral drugs for Covid-19-interim WHO solidarity trial results, New England journal of medicine
Coomes, Haghbayan, Interleukin-6 in COVID-19: a systematic review and meta-analysis, Reviews in medical virology
Davies, Adlimoghaddam, Albensi, The effect of COVID-19 on NF-κB and neurological manifestations of disease, Molecular Neurobiology
Davis, Comparative toxicogenomics database (CTD): update 2021, Nucleic acids research
De Flora, Balansky, La Maestra, Antioxidants and COVID-19, Journal of preventive medicine and hygiene
Del Toro, The IntAct database: efficient access to fine-grained molecular interaction data, Nucleic acids research
Diaz, Remdesivir and mortality in patients with coronavirus disease 2019, Clinical Infectious Diseases
Dong, Du, Gardner, An interactive web-based dashboard to track COVID-19 in real time, The Lancet infectious diseases
Dryden-Peterson, Nirmatrelvir plus ritonavir for early COVID-19 in a large US health system: a population-based cohort study, Annals of internal medicine
Durante, Glutamine Deficiency Promotes Immune and Endothelial Cell Dysfunction in COVID-19, International Journal of Molecular Sciences
El Bairi, Repurposing anticancer drugs for the management of COVID-19, European Journal of Cancer
El-Tanani, Phase II, Double-Blinded, Randomized, Placebo-Controlled Clinical Trial Investigating the Efficacy of Mebendazole in the Management of Symptomatic COVID-19 Patients, Pharmaceuticals
Ennis, Tiligada, Histamine receptors and COVID-19, Inflammation Research
Eriksson, Combining hypothesis-and data-driven neuroscience modeling in FAIR workflows, Elife
Ferdinands, Waning of vaccine effectiveness against moderate and severe covid-19 among adults in the US from the VISION network: test negative, casecontrol study
Freshour, Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts, Nucleic acids research
Gajjela, Zhou, Calming the cytokine storm of COVID-19 through inhibition of JAK2/STAT3 signaling, Drug discovery today
Giamarellos-Bourboulis, Complex immune dysregulation in COVID-19 patients with severe respiratory failure, Cell host & microbe
Gordon, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Group, Aspirin in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial, Lancet
Group, Dexamethasone in hospitalized patients with Covid-19, New England Journal of Medicine
Group, Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial, Lancet
Gupta, Association between early treatment with tocilizumab and mortality among critically ill patients with COVID-19
Gupta, Association between early treatment with tocilizumab and mortality among critically ill patients with COVID-19, JAMA internal medicine
Guy, Rapid repurposing of drugs for COVID-19, Science
Halpin, A prospective, single-center, randomized phase 2 trial of etoposide in severe COVID-19
Hamizi, Aouidane, Belaaloui, Etoposide-based therapy for severe forms of COVID-19, Medical Hypotheses
Hammond, Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19, New England Journal of Medicine
Han, Discovery of podofilox as a potent cGAMP-STING signaling enhancer with antitumor activity, Cancer Immunology Research
Heskin, Caution required with use of ritonavir-boosted PF-07321332 in COVID-19 management, The Lancet
Himmelstein, Systematic integration of biomedical knowledge prioritizes drugs for repurposing, Elife
Hinkson, COVID-19 Treatment Guidelines
Horby, Effect of dexamethasone in hospitalized patients with COVID-19preliminary report, MedRxiv
Hsieh, Drug repurposing for COVID-19 using graph neural network and harmonizing multiple evidence, Scientific reports
Hudzik, Nowak, Zubelewicz-Szkodzinska, Consideration of immunomodulatory actions of morphine in COVID-19-Short report, Eur Rev Med Pharmacol Sci
Ioannidis, Drkg-drug repurposing knowledge graph for covid-19
Izquierdo-Alonso, N-acetylcysteine for prevention and treatment of COVID-19: Current state of evidence and future directions, Journal of infection and public health
Jamal, Alharbi, Ahmad, Identification of doxorubicin as a potential therapeutic against SARS-CoV-2 (COVID-19) protease: a molecular docking and dynamics simulation studies, Journal of Biomolecular Structure and Dynamics
Jennings, Parks, Curcumin as an antiviral agent, Viruses
Johnson, Etoposide selectively ablates activated T cells to control the immunoregulatory disorder hemophagocytic lymphohistiocytosis, The Journal of Immunology
Jones, Hunter, Is IL-6 a key cytokine target for therapy in COVID-19?, Nature Reviews Immunology
Jung, Random walk with restart on large graphs using block elimination, ACM Transactions on Database Systems (TODS)
Jørgensen, Sirolimus interferes with the innate response to bacterial products in human whole blood by attenuation of IL-10 production, Scandinavian journal of immunology
Karsulovic, mTORC inhibitor Sirolimus deprograms monocytes in "cytokine storm" in SARS-CoV2 secondary hemophagocytic lymphohistiocytosis-like syndrome, Clinical Immunology
Khaledi, COVID-19 and the potential of Janus family kinase (JAK) pathway inhibition: A novel treatment strategy, Frontiers in medicine
Khalili, Novel coronavirus treatment with ribavirin: groundwork for an evaluation concerning COVID-19, Journal of medical virology
Kim, Serious Clinical Outcomes of COVID-19 Related to Acetaminophen or NSAIDs from a Nationwide Population-Based Cohort Study, International Journal of Environmental Research and Public Health
Kim, The architecture of SARS-CoV-2 transcriptome, Cell
Kocks, A potential harmful effect of dexamethasone in non-severe COVID-19: results from the COPPER-pilot study, ERJ Open Research
Laforge, Tissue damage from neutrophil-induced oxidative stress in COVID-19, Nature Reviews Immunology
Laing, A dynamic COVID-19 immune signature includes associations with poor prognosis, Nature medicine
Lau, Real-world COVID-19 vaccine effectiveness against the Omicron BA. 2 variant in a SARS-CoV-2 infection-naive population, Nature medicine
Li, Tenofovir disoproxil fumarate and coronavirus disease 2019 outcomes in men with HIV, Aids
Liew, SARS-CoV-2 neutralizing antibody bebtelovimab-a systematic scoping review and meta-analysis, Frontiers in Immunology
Liu, DrugCombDB: a comprehensive database of drug combinations toward the discovery of combinatorial therapy, Nucleic acids research
Lucas, Longitudinal analyses reveal immunological misfiring in severe COVID-19, Nature
Lv, Efficient processing node proximity via random walk with restart
Ma, Does aspirin have an effect on risk of death in patients with COVID-19? A meta-analysis, European Journal of Clinical Pharmacology
Ma, Homo-harringtonine, highly effective against coronaviruses, is safe in treating COVID-19 by nebulization, Science China Life Sciences
Mahdian, Ebrahim-Habibi, Zarrabi, Drug repurposing using computational methods to identify therapeutic options for COVID-19, Journal of Diabetes & Metabolic Disorders
Malone, COVID-19: famotidine, histamine, mast cells, and mechanisms, Frontiers in Pharmacology
Manjani, Effects of acetaminophen on outcomes in patients hospitalized with COVID-19, Chest
Mashauri, Covid-19 Histamine theory: Why antihistamines should be incorporated as the basic component in Covid-19 management?, Health Science Reports
Menni, COVID-19 vaccine waning and effectiveness and side-effects of boosters: a prospective community study from the ZOE COVID Study, The Lancet Infectious Diseases
Mirjalili, Does Losartan reduce the severity of COVID-19 in hypertensive patients?, BMC Cardiovascular Disorders
Mushtaq, Tocilizumab in critically ill COVID-19 patients: An observational study, International Immunopharmacology
Nair, Sharma, Tiwary, Glutathione deficiency in COVID19 illnessdoes supplementation help?, Saudi Journal of Anaesthesia
Nevalainen, Effect of remdesivir post hospitalization for COVID-19 infection from the randomized SOLIDARITY Finland trial, Nature Communications
Nguyen, Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses, Science Advances
Organization, The use of non-steroidal anti-inflammatory drugs (NSAIDs) in patients with COVID-19
Overmyer, Large-scale multi-omic analysis of COVID-19 severity, Cell systems
Panda, Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV2 Infection
Pashmforosh, Possible Benefits of Paclitaxel Therapy for COVID-19, Pharmaceutical and Biomedical Research
Patel, Azithromycin for mild-to-moderate COVID-19, The Lancet Respiratory Medicine
Pathania, Fenizia, Cyclosporine a inhibits viral infection and release as well as cytokine production in lung cells by three SARS-CoV-2 variants, Microbiology Spectrum
Patocka, Rapamycin: drug repurposing in SARS-CoV-2 infection, Pharmaceuticals
Pektaş, Gürsoy, Demirbilek, The use of pregabalin in Intensive Care Unit in the treatment of Covid-19-related pain and cough
Perreau, The cytokines HGF and CXCL13 predict the severity and the mortality in COVID-19 patients, Nature Communications
Puskarich, Efficacy of losartan in hospitalized patients with COVID-19induced lung injury: a randomized clinical trial, JAMA Network Open
Páez-Franco, Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients, Scientific reports
Ravichandran, An open label randomized clinical trial of Indomethacin for mild and moderate hospitalised Covid-19 patients, Scientific reports
Ravid, Leiva, Chitalia, Janus kinase signaling pathway and its role in COVID-19 inflammatory, vascular, and thrombotic manifestations, Cells
Rheingold, Zinc Supplementation Associated With a Decrease in Mortality in COVID-19 Patients: A Meta-Analysis, Cureus
Rincon-Arevalo, Altered increase in STAT1 expression and phosphorylation in severe COVID-19, European Journal of Immunology
Ripamonti, HDAC inhibition as potential therapeutic strategy to restore the deregulated immune response in severe COVID-19, Frontiers in immunology
Sajgure, Safety and efficacy of mycophenolate in COVID-19: A nonrandomised prospective study in western India, The Lancet Regional Health-Southeast Asia
Salerni, Vinblastine induces acute, cell cycle phase-independent apoptosis in some leukemias and lymphomas and can induce acute apoptosis in others when Mcl-1 is suppressed, Molecular cancer therapeutics
Salt, Palmer, Exploiting the anti-inflammatory effects of AMP-activated protein kinase activation, Expert opinion on investigational drugs
Samaee, Tocilizumab for treatment patients with COVID-19: recommended medication for novel disease, International immunopharmacology
Samuel, Varghese, Büsselberg, Therapeutic potential of metformin in COVID-19: reasoning for its protective role, Trends in microbiology
Schoot, Immunosuppressive drugs and COVID-19: a review, Frontiers in pharmacology
Schwartz, Does ivermectin have a place in the treatment of mild Covid-19? New microbes and new infections
Singh, A comparative study of 5-fluorouracil, doxorubicin, methotrexate, paclitaxel for their inhibition ability for Mpro of nCoV: Molecular docking and molecular dynamics simulations, Journal of the Indian Chemical Society
Sivapalasingam, Efficacy and safety of sarilumab in hospitalized patients with coronavirus disease 2019: a randomized clinical trial, Clinical Infectious Diseases
Sosa, A literature-based knowledge graph embedding method for identifying drug repurposing opportunities in rare diseases
Sperry, Target-agnostic drug prediction integrated with medical record analysis uncovers differential associations of statins with increased survival in COVID-19 patients, PLOS Computational Biology
Sterne, Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis, Jama
Su, Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19, Cell
Suresh, Therapeutic potential of curcumin in ARDS and COVID-19, Clinical and Experimental Pharmacology and Physiology
Suriawinata, Mehta, Iron and iron-related proteins in COVID-19, Clinical and Experimental Medicine
Szklarczyk, The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets, Nucleic acids research
Takimoto, Anticancer drug development at the US National Cancer Institute, Cancer chemotherapy and pharmacology
Tanaka, Narazaki, Kishimoto, Interleukin (IL-6) immunotherapy, Cold Spring Harbor perspectives in biology
Teixeira, Simvastatin downregulates the SARS-CoV-2-induced inflammatory response and impairs viral infection through disruption of lipid rafts, Frontiers in Immunology
Telenti, After the pandemic: perspectives on the future trajectory of COVID-19, Nature
Thangam, The role of histamine and histamine receptors in mast cellmediated allergy and inflammation: the hunt for new therapeutic targets, Frontiers in immunology
Tomazou, Multi-omics data integration and network-based analysis drives a multiplex drug repurposing approach to a shortlist of candidate drugs against COVID-19, Briefings in bioinformatics
Vahedian-Azimi, Effectiveness of curcumin on outcomes of hospitalized COVID-19 patients: A systematic review of clinical trials, Nutrients
Valle, An inflammatory cytokine signature predicts COVID-19 severity and survival, Nature medicine
Vitiello, Porta, Ferrara, Correlation between the use of statins and COVID-19: what do we know?, BMJ Evidence-Based Medicine
Wang, Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study, Journal of chemical information and modeling
Wang, Vinblastine resets tumor-associated macrophages toward M1 phenotype and promotes antitumor immune response, Journal for Immunotherapy of Cancer
Weisberg, Repurposing of kinase inhibitors for treatment of COVID-19, Pharmaceutical research
Wishart, DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic acids research
Wróblewska, The Role of Glutathione in Selected Viral Diseases, Antioxidants
Wu, The SARS-CoV-2 induced targeted amino acid profiling in patients at hospitalized and convalescent stage, Bioscience Reports
Xiu, Fludarabine inhibits type I interferon-induced expression of the SARS-CoV-2 receptor angiotensin-converting enzyme 2, Cellular & Molecular Immunology
Xu, Pathological findings of COVID-19 associated with acute respiratory distress syndrome, The Lancet respiratory medicine
Xu, Ribavirin treatment for critically ill COVID-19 patients: An observational study, Infection and Drug Resistance
Zarkovic, Post-mortem findings of inflammatory cells and the association of 4-hydroxynonenal with systemic vascular and oxidative stress in lethal COVID-19, Cells
Zhang, Immune evasive effects of SARS-CoV-2 variants to COVID-19 emergency used vaccines, Frontiers in Immunology
Zhou, Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study, The lancet
Zhou, Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2, Cell discovery
Zhou, Role of AMP-activated protein kinase in mechanism of metformin action, The Journal of clinical investigation
Zhou, The role of SARS-CoV-2-mediated NF-κB activation in COVID-19 patients, Hypertension Research
Žarković, The impact of severe COVID-19 on plasma antioxidants, Molecules
DOI record:
{
"DOI": "10.58647/drugarxiv.pr000010.v1",
"URL": "http://dx.doi.org/10.58647/DRUGARXIV.PR000010.v1",
"abstract": "<jats:p> \n <jats:bold>Background:</jats:bold>The development and roll-out of vaccines, and the use of various drugs have contributed to controlling the COVID-19 pandemic. Nevertheless, challenges such as the inequitable distribution of vaccines, the influence of emerging viral lineages and immune evasive variants on vaccine efficacy, and the inadequate immune defense in subgroups of the population continue to motivate the development of new drugs to combat the disease.</jats:p>\n <jats:p> \n <jats:bold>Aim:</jats:bold>In this study, we sought to identify, prioritize, and characterize drug repurposing candidates appropriate for treating mild, moderate, or severe COVID-19 using a network-based integrative approach that systematically integrates drug-related data and multi-omics datasets.</jats:p>\n <jats:p> \n <jats:bold>Methods</jats:bold>: We leveraged drug data, and multi-omics data, and used a random walk restart algorithm to explore an integrated knowledge graph comprised of three sub-graphs: (i) a COVID-19 knowledge graph, (ii) a drug repurposing knowledge graph, and (iii) a COVID-19 disease-state specific omics graph.</jats:p>\n <jats:p> \n <jats:bold>Results:</jats:bold>We prioritized twenty FDA-approved agents as potential candidate drugs for mild, moderate, and severe COVID-19 disease phases. Specifically, drugs that could stimulate immune cell recruitment and activation including histamine, curcumin, and paclitaxel have potential utility in mild disease states to mitigate disease progression. Drugs like omacetaxine, crizotinib, and vorinostat that exhibit antiviral properties and have the potential to inhibit viral replication can be considered for mild to moderate COVID-19 disease states. Also, given the association between antioxidant deficiency and high inflammatory factors that trigger cytokine storms, antioxidants like glutathione can be considered for moderate disease states. Drugs that exhibit potent anti-inflammatory effects like (i) anti-inflammatory drugs (sarilumab and tocilizumab), (ii) corticosteroids (dexamethasone and hydrocortisone), and (iii) immunosuppressives (sirolimus and cyclosporine) are potential candidates for moderate to severe disease states that trigger a hyperinflammatory cascade of COVID-19.</jats:p>\n <jats:p> \n <jats:bold>Conclusion:</jats:bold>Our study demonstrates that the multi-omics data-driven integrative analysis within the drug data enables prioritizing drug candidates for COVID-19 disease phases, offering a comprehensive basis for therapeutic strategies that can be brought to market quickly given their established safety profiles. Importantly, the multi-omics data-driven integrative analysis within the drug data approach implemented here can be used to prioritize drug repurposing candidates appropriate for other diseases.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0002-2980-1392",
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/03p74gp79",
"id-type": "ROR"
}
],
"name": "Computational Biology Division, Department of Integrative Biomedical Sciences, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa"
}
],
"authenticated-orcid": false,
"family": "Agamah",
"given": "Francis Edem",
"sequence": "first"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/05wg1m734",
"id-type": "ROR"
}
],
"name": "Department of Medical BioSciences, Radboud University Medical Center Nijmegen, The Netherlands"
}
],
"family": "Ederveen",
"given": "Thomas H.A.",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/03p74gp79",
"id-type": "ROR"
}
],
"name": "Computational Biology Division, Department of Integrative Biomedical Sciences, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa"
}
],
"family": "Skelton",
"given": "Michelle",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/03p74gp79",
"id-type": "ROR"
}
],
"name": "Computational Biology Division, Department of Integrative Biomedical Sciences, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa"
}
],
"family": "Martin",
"given": "Darren P.",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/049e6bc10",
"id-type": "ROR"
}
],
"name": "Department of Applied Science, Faculty of Health and Life Sciences, Northumbria University, Newcastle, Tyne and Wear, NE1 8ST, UK"
}
],
"family": "Chimusa",
"given": "Emile R.",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/05wg1m734",
"id-type": "ROR"
}
],
"name": "Department of Medical BioSciences, Radboud University Medical Center Nijmegen, The Netherlands"
}
],
"family": "'t Hoen",
"given": "Peter A.C. 't Hoen",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
4,
17
]
],
"date-time": "2024-04-17T13:50:17Z",
"timestamp": 1713361817000
},
"deposited": {
"date-parts": [
[
2024,
4,
17
]
],
"date-time": "2024-04-17T13:50:18Z",
"timestamp": 1713361818000
},
"funder": [
{
"award": [
"184.034.019"
],
"name": "Dutch Organization of Scientific Research"
},
{
"award": [
"871096"
],
"name": "European Union to the EATRIS-Plus infrastructure project"
}
],
"indexed": {
"date-parts": [
[
2024,
4,
18
]
],
"date-time": "2024-04-18T02:13:26Z",
"timestamp": 1713406406464
},
"institution": [
{
"name": "ScienceOpen"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
4,
16
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
4,
16
]
],
"date-time": "2024-04-16T00:00:00Z",
"timestamp": 1713225600000
}
}
],
"link": [
{
"URL": "https://drugrepocentral.scienceopen.com/hosted-document?doi=10.58647/DRUGARXIV.PR000010.v1",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "5403",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
4,
16
]
]
},
"prefix": "10.58647",
"published": {
"date-parts": [
[
2024,
4,
16
]
]
},
"publisher": "ScienceOpen",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://drugrepocentral.scienceopen.com/hosted-document?doi=10.58647/DRUGARXIV.PR000010.v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases",
"type": "posted-content"
}