Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach
et al., Future Medicine, doi:10.2217/fvl-2020-0342, Mar 2021
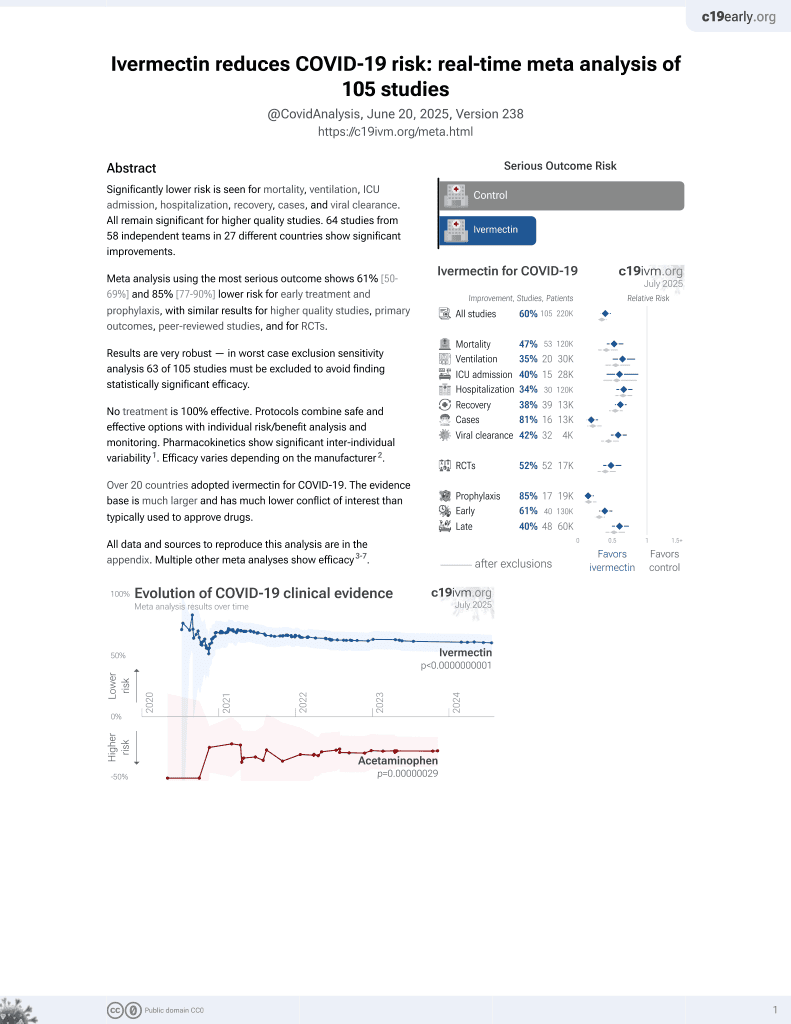
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico analysis finding that ivermectin has high binding affinity for the SARS-CoV-2 viral spike protein, main protease, replicase, and human TMPRSS2 receptors.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Choudhury et al., 25 Mar 2021, peer-reviewed, 7 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach
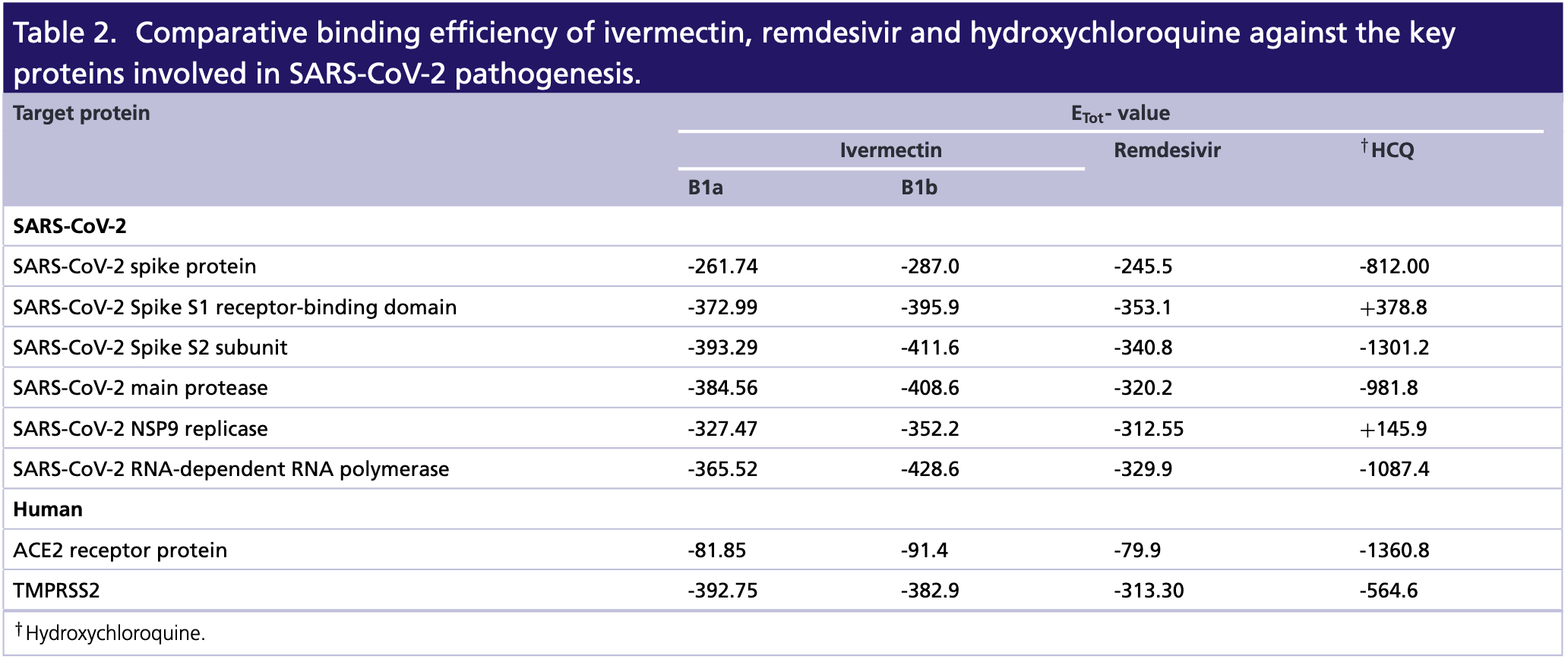
Aim: COVID-19 is currently the biggest threat to mankind. Recently, ivermectin (a US FDA-approved antiparasitic drug) has been explored as an anti-SARS-CoV-2 agent. Herein, we have studied the possible mechanism of action of ivermectin using in silico approaches. Materials & methods: Interaction of ivermectin against the key proteins involved in SARS-CoV-2 pathogenesis were investigated through molecular docking and molecular dynamic simulation. Results: Ivermectin was found as a blocker of viral replicase, protease and human TMPRSS2, which could be the biophysical basis behind its antiviral efficiency. The antiviral action and ADMET profile of ivermectin was on par with the currently used anticorona drugs such as hydroxychloroquine and remdesivir. Conclusion: Our study enlightens the candidature of ivermectin as an effective drug for treating COVID-19.
has relatively much higher water solubility and lipophilicity, further, having lesser skin permeation on the other hand (Supplementary Table 2 ). The three drugs included in the study are FDA-approved drugs and used for treating various parasitic (ivermectin and hydroxychloroquine) and viral (remdesivir) infections of human. However, to present the suitability of ivermectin for treating COVID-19, we have compared the pharmacological properties of ivermectin with the other two drugs. Taken together, our data on the interaction between ivermectin and viral proteins indicated that ivermectin majorly acts by interfering with the viral entry through inhibiting the function of spike protein and protease. These studies also indicate that ivermectin may also target human ACE2 and TMPRSS2 for exerting its inhibitory action over SARS-CoV-2. However, all these in silico studies require subsequent experimental validation, which could enable Ivermectin as a drug of reliance to be used for counteracting the viral growth.
Conclusion Developing an effective therapeutic against COVID-19 is currently the utmost interest to the scientific communities. The present study depicts comparative binding efficacy of a promising FDA-approved drug, ivermectin, against major pathogenic proteins of SARS-CoV-2 and their human counterparts involved in host-pathogen interaction. Herein, our in silico data have indicated that ivermectin efficiently utilizes viral spike protein, main protease, replicase and..
References
Beigel, Nam, Adams, Advances in respiratory virus therapeutics -a meeting report from the 6th ISIRV Antiviral Group conference, Antiviral Res
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res
Cao, COVID-19: immunopathology and its implications for therapy, Nat. Rev. Immunol
Chaccour, Kobylinski, Bassat, Ivermectin to reduce malaria transmission: a research agenda for a promising new tool for elimination, Malar. J
Choudhury, Das, Patra, Mukherjee, In silico analyses on the comparative sensing of SARS-CoV-2 mRNA by the intracellular TLRs of humans, J. Med. Virol
Choudhury, Mukherjee, In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs, J. Med. Virol
Eweas, Alhossary, As, Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2, Front. Microbiol
Fantini, Scala, Chahinian, Yahi, Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection, Int. J. Antimicrob. Agents
Fda, Merck, Annual Highlights Archives -Mectizan Donation Program
Formiga, Leblanc, Souza Rebouc ¸as, Farias, Oliveira et al., Ivermectin: an award-winning drug with expected antiviral activity against COVID-19, J. Control. Rel
Gautret, Lagier, Parola, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int. J. Antimicrob. Agents
Gordon, Tchesnokov, Woolner, Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency, J. Biol. Chem
Heidary, Gharebaghi, Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen, J. Antibiot. (Tokyo)
Kihara, Chen, David, Quality assessment of protein structure models, Curr. Protein Pept. Sci
Laing, Devaney, Ivermectin -old drug, new tricks?, Trends Parasitol
Lau, Khosrawipour, Kocbach, The association between international and domestic air traffic and the coronavirus (COVID-19) outbreak, J. Microbiol. Immunol. Infect
Lehrer, Rheinstein, Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2, vivo (Athens, Greece)
Li, Zhao, Zhan, Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cell. Physiol
López-Blanco, Aliaga, Quintana-Ortí, Chacón, iMODS: internal coordinates normal mode analysis server, Nucleic Acids Res
Mason, Pathogenesis of COVID-19 from a cell biology perspective, Eur. Respir. J
Mousavizadeh, Ghasemi, Genotype and phenotype of COVID-19: their roles in pathogenesis, J. Microbiol. Immunol. Infect
Mukherjee, Mukherjee, Gayen, Roy, Babu, Metabolic inhibitors as antiparasitic drugs: pharmacological, biochemical and molecular perspectives, Curr. Drug Metab
Nicolas, Maia, Bassat, Safety of oral ivermectin during pregnancy: a systematic review and meta-analysis, Lancet. Glob. Heal
Ong, Tan, Chia, Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient, JAMA
Patra, Das, Mukherjee, Targeting human TLRs to combat COVID-19: a solution?, J. Med. Virol
Peng, Xu, Li, Cheng, Zhou et al., Transmission routes of 2019-nCoV and controls in dental practice, Int. J. Oral Sci
Rizzo, Ivermectin, antiviral properties and COVID-19: a possible new mechanism of action, Naunyn-Schmiedebergs Arch. Pharmacol
Rothan, Byrareddy, The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak, J. Autoimmun
Swargiary, Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1
Thomsen, Sanuku, Baea, Efficacy, safety, and pharmacokinetics of coadministered diethylcarbamazine, albendazole, and ivermectin for treatment of Bancroftian Filariasis, Clin. Infect. Dis
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell
Wan, Shang, Graham, Baric, Li, Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus, J. Virol
Who, WHO Coronavirus Disease (COVID-19) Situation Reports
Yi, Lagniton, Ye, Li, Xu, COVID-19: what has been learned and to be learned about the novel coronavirus disease, Int. J. Biol. Sci
DOI record:
{
"DOI": "10.2217/fvl-2020-0342",
"ISSN": [
"1746-0794",
"1746-0808"
],
"URL": "http://dx.doi.org/10.2217/fvl-2020-0342",
"abstract": "<jats:p> Aim: COVID-19 is currently the biggest threat to mankind. Recently, ivermectin (a US FDA-approved antiparasitic drug) has been explored as an anti-SARS-CoV-2 agent. Herein, we have studied the possible mechanism of action of ivermectin using in silico approaches. Materials & methods: Interaction of ivermectin against the key proteins involved in SARS-CoV-2 pathogenesis were investigated through molecular docking and molecular dynamic simulation. Results: Ivermectin was found as a blocker of viral replicase, protease and human TMPRSS2, which could be the biophysical basis behind its antiviral efficiency. The antiviral action and ADMET profile of ivermectin was on par with the currently used anticorona drugs such as hydroxychloroquine and remdesivir. Conclusion: Our study enlightens the candidature of ivermectin as an effective drug for treating COVID-19. </jats:p>",
"alternative-id": [
"10.2217/fvl-2020-0342"
],
"author": [
{
"affiliation": [
{
"name": "Integrative Biochemistry & Immunology Laboratory, Department of Animal Science, Kazi Nazrul University, Asansol 713340, West Bengal, India"
}
],
"family": "Choudhury",
"given": "Abhigyan",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Integrative Biochemistry & Immunology Laboratory, Department of Animal Science, Kazi Nazrul University, Asansol 713340, West Bengal, India"
}
],
"family": "Das",
"given": "Nabarun C",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Integrative Biochemistry & Immunology Laboratory, Department of Animal Science, Kazi Nazrul University, Asansol 713340, West Bengal, India"
}
],
"family": "Patra",
"given": "Ritwik",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Zoology, Fakir Mohan University, Balasore 756020, Odisha, India"
}
],
"family": "Bhattacharya",
"given": "Manojit",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Zoology, Vidyasagar University, Midnapore 721102, West Bengal, India"
}
],
"family": "Ghosh",
"given": "Pratik",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Zoology, Vidyasagar University, Midnapore 721102, West Bengal, India"
}
],
"family": "Patra",
"given": "Bidhan C",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5709-9190",
"affiliation": [
{
"name": "Integrative Biochemistry & Immunology Laboratory, Department of Animal Science, Kazi Nazrul University, Asansol 713340, West Bengal, India"
}
],
"authenticated-orcid": false,
"family": "Mukherjee",
"given": "Suprabhat",
"sequence": "additional"
}
],
"container-title": "Future Virology",
"container-title-short": "Future Virology",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
3,
25
]
],
"date-time": "2021-03-25T11:21:50Z",
"timestamp": 1616671310000
},
"deposited": {
"date-parts": [
[
2021,
12,
21
]
],
"date-time": "2021-12-21T14:13:25Z",
"timestamp": 1640096005000
},
"indexed": {
"date-parts": [
[
2024,
5,
11
]
],
"date-time": "2024-05-11T15:15:14Z",
"timestamp": 1715440514486
},
"is-referenced-by-count": 46,
"issue": "4",
"issued": {
"date-parts": [
[
2021,
4
]
]
},
"journal-issue": {
"issue": "4",
"published-print": {
"date-parts": [
[
2021,
4
]
]
}
},
"language": "en",
"link": [
{
"URL": "https://www.futuremedicine.com/doi/pdf/10.2217/fvl-2020-0342",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1057",
"original-title": [],
"page": "277-291",
"prefix": "10.2217",
"published": {
"date-parts": [
[
2021,
4
]
]
},
"published-print": {
"date-parts": [
[
2021,
4
]
]
},
"publisher": "Future Medicine Ltd",
"reference": [
{
"DOI": "10.1016/j.jaut.2020.102433",
"doi-asserted-by": "publisher",
"key": "B1"
},
{
"key": "B2",
"unstructured": "WHO. WHO Coronavirus Disease (COVID-19) Situation Reports-209. (2020). www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports"
},
{
"author": "Mousavizadeh L",
"first-page": "30082",
"issue": "20",
"journal-title": "J. Microbiol. Immunol. Infect.",
"key": "B3",
"volume": "1182",
"year": "2020"
},
{
"DOI": "10.1038/s41368-020-0075-9",
"doi-asserted-by": "publisher",
"key": "B4"
},
{
"DOI": "10.1001/jama.2020.3227",
"doi-asserted-by": "publisher",
"key": "B5"
},
{
"DOI": "10.1016/j.jmii.2020.03.026",
"doi-asserted-by": "publisher",
"key": "B6"
},
{
"DOI": "10.1183/13993003.00607-2020",
"doi-asserted-by": "publisher",
"key": "B7"
},
{
"DOI": "10.1128/JVI.00127-20",
"doi-asserted-by": "publisher",
"key": "B8"
},
{
"DOI": "10.1038/s41577-020-0308-3",
"doi-asserted-by": "publisher",
"key": "B9"
},
{
"DOI": "10.7150/ijbs.45134",
"doi-asserted-by": "publisher",
"key": "B10"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105949",
"doi-asserted-by": "publisher",
"key": "B11"
},
{
"DOI": "10.1016/j.antiviral.2019.04.006",
"doi-asserted-by": "publisher",
"key": "B12"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "B13"
},
{
"DOI": "10.1016/j.pt.2017.02.004",
"doi-asserted-by": "publisher",
"key": "B14"
},
{
"DOI": "10.1038/s41429-020-0336-z",
"doi-asserted-by": "publisher",
"key": "B15"
},
{
"DOI": "10.1093/nar/gku339",
"doi-asserted-by": "publisher",
"key": "B16"
},
{
"DOI": "10.2174/138920309788452173",
"doi-asserted-by": "publisher",
"key": "B17"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"doi-asserted-by": "publisher",
"key": "B18"
},
{
"DOI": "10.1002/jmv.25987",
"doi-asserted-by": "publisher",
"key": "B19"
},
{
"key": "B20",
"unstructured": "FDA. Merck STROMECTOL ® (IVERMECTIN). (2009). www.accessdata.fda.gov/drugsatfda_docs/label/2009/050742s026lbl.pdf"
},
{
"key": "B21",
"unstructured": "Mectizan. Annual Highlights Archives – Mectizan Donation Program. (2017). https://mectizan.org/news-resources/hub-cat/annual-highlights/"
},
{
"DOI": "10.1093/cid/civ882",
"doi-asserted-by": "publisher",
"key": "B22"
},
{
"DOI": "10.2174/1389200217666161004143152",
"doi-asserted-by": "publisher",
"key": "B23"
},
{
"DOI": "10.1186/1475-2875-12-153",
"doi-asserted-by": "publisher",
"key": "B24"
},
{
"DOI": "10.1016/S2214-109X(19)30453-X",
"doi-asserted-by": "publisher",
"key": "B25"
},
{
"DOI": "10.1007/s00210-020-01902-5",
"doi-asserted-by": "publisher",
"key": "B26"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105960",
"doi-asserted-by": "publisher",
"key": "B27"
},
{
"DOI": "10.1074/jbc.RA120.013679",
"doi-asserted-by": "publisher",
"key": "B28"
},
{
"author": "Swargiary A",
"journal-title": "Research Square",
"key": "B29",
"year": "2020"
},
{
"DOI": "10.3389/fmicb.2020.592908",
"doi-asserted-by": "publisher",
"key": "B30"
},
{
"author": "Lehrer S",
"first-page": "3023",
"issue": "5",
"journal-title": "In vivo (Athens, Greece)",
"key": "B31",
"volume": "34",
"year": "2020"
},
{
"DOI": "10.1002/jcp.30055",
"doi-asserted-by": "publisher",
"key": "B32"
},
{
"DOI": "10.1016/j.jconrel.2020.10.009",
"doi-asserted-by": "publisher",
"key": "B33"
},
{
"DOI": "10.1002/jmv.26387",
"doi-asserted-by": "publisher",
"key": "B34"
},
{
"DOI": "10.1002/jmv.26776",
"doi-asserted-by": "publisher",
"key": "B35"
}
],
"reference-count": 35,
"references-count": 35,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.futuremedicine.com/doi/10.2217/fvl-2020-0342"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an <i>in silico</i> approach",
"type": "journal-article",
"volume": "16"
}