Prioritization of Anti-SARS-Cov-2 Drug Repurposing Opportunities Based on Plasma and Target Site Concentrations Derived from their Established Human Pharmacokinetics
et al., Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1909, May 2020
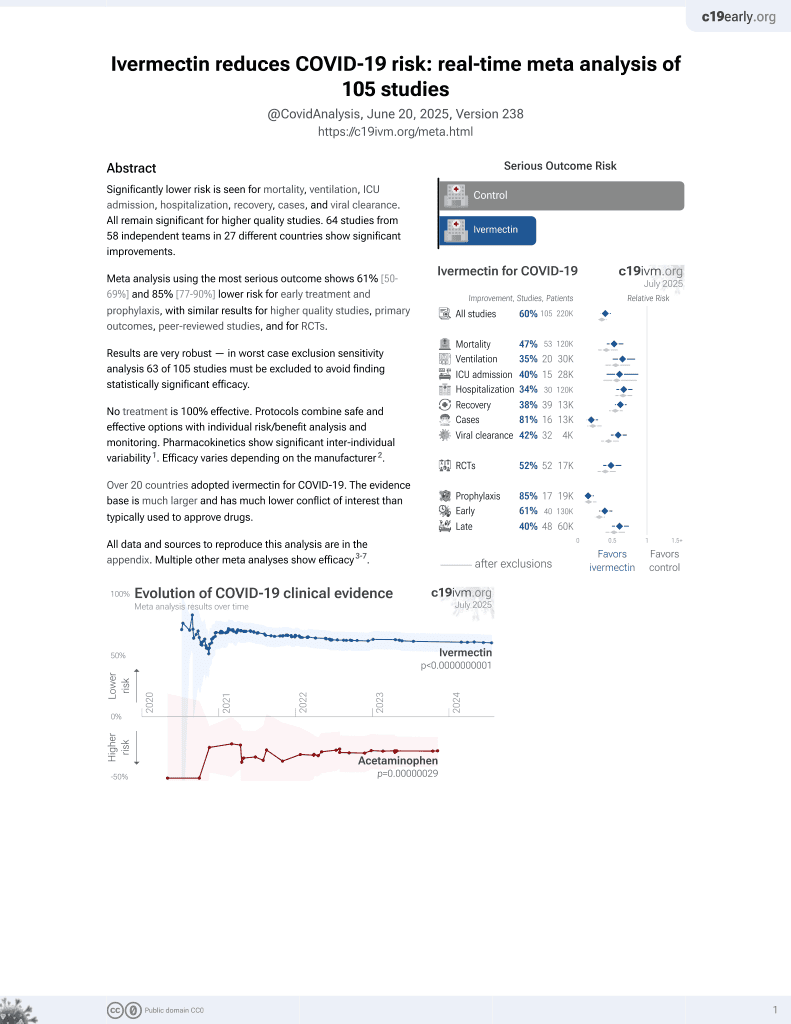
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
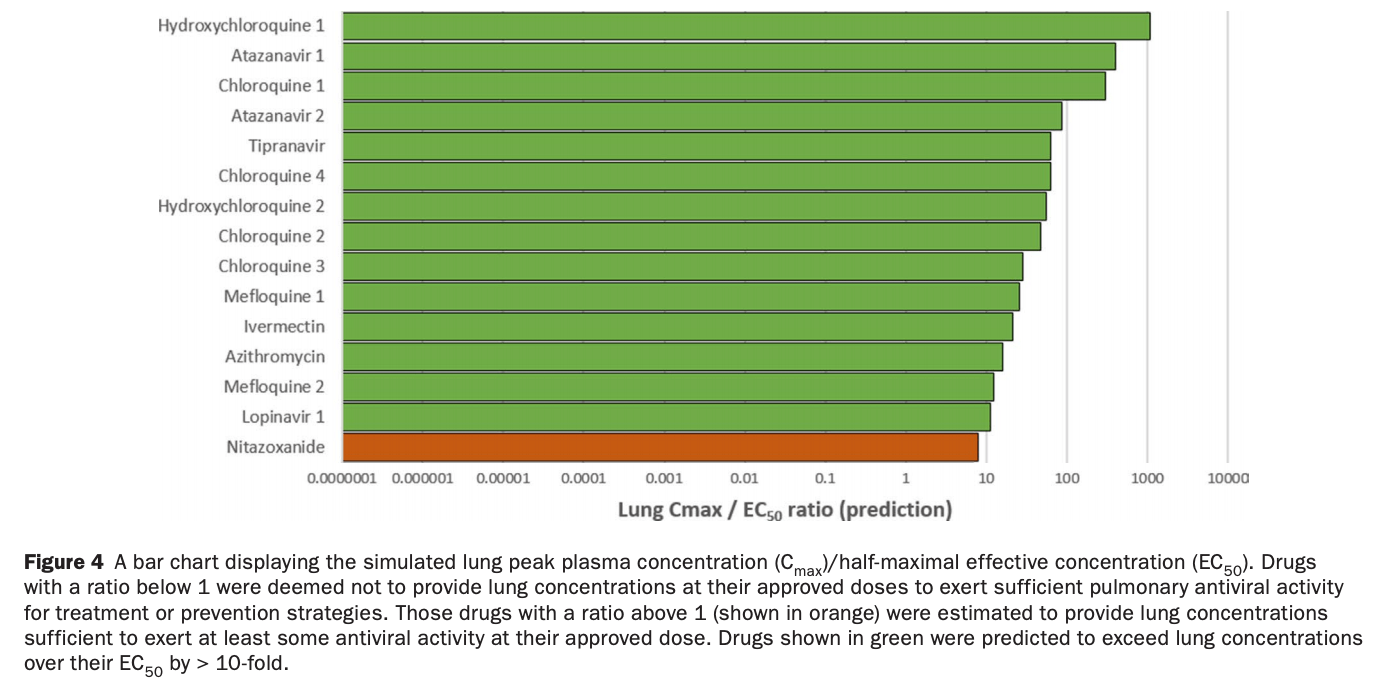
Pharmacokinetic analysis predicting that ivermectin, HCQ, CQ, and azithromycin will achieve lung concentrations well over 10 times higher than the reported EC50. Nitazoxanide had a lung tissue Cmax/EC50 of 7.8.
74 preclinical studies support the efficacy of ivermectin for COVID-19:
Ivermectin, better known for antiparasitic activity, is a broad spectrum antiviral with activity against many viruses including H7N771, Dengue37,72,73 , HIV-173, Simian virus 4074, Zika37,75,76 , West Nile76, Yellow Fever77,78, Japanese encephalitis77, Chikungunya78, Semliki Forest virus78, Human papillomavirus57, Epstein-Barr57, BK Polyomavirus79, and Sindbis virus78.
Ivermectin inhibits importin-α/β-dependent nuclear import of viral proteins71,73,74,80 , shows spike-ACE2 disruption at 1nM with microfluidic diffusional sizing38, binds to glycan sites on the SARS-CoV-2 spike protein preventing interaction with blood and epithelial cells and inhibiting hemagglutination41,81, shows dose-dependent inhibition of wildtype and omicron variants36, exhibits dose-dependent inhibition of lung injury61,66, may inhibit SARS-CoV-2 via IMPase inhibition37, may inhibit SARS-CoV-2 induced formation of fibrin clots resistant to degradation9, inhibits SARS-CoV-2 3CLpro54, may inhibit SARS-CoV-2 RdRp activity28, may minimize viral myocarditis by inhibiting NF-κB/p65-mediated inflammation in macrophages60, may be beneficial for COVID-19 ARDS by blocking GSDMD and NET formation82, may interfere with SARS-CoV-2's immune evasion via ORF8 binding4, may inhibit SARS-CoV-2 by disrupting CD147 interaction83-86, may inhibit SARS-CoV-2 attachment to lipid rafts via spike NTD binding2, shows protection against inflammation, cytokine storm, and mortality in an LPS mouse model sharing key pathological features of severe COVID-1959,87, may be beneficial in severe COVID-19 by binding IGF1 to inhibit the promotion of inflammation, fibrosis, and cell proliferation that leads to lung damage8, may minimize SARS-CoV-2 induced cardiac damage40,48, may counter immune evasion by inhibiting NSP15-TBK1/KPNA1 interaction and restoring IRF3 activation88, may disrupt SARS-CoV-2 N and ORF6 protein nuclear transport and their suppression of host interferon responses1, reduces TAZ/YAP nuclear import, relieving SARS-CoV-2-driven suppression of IRF3 and NF-κB antiviral pathways35, increases Bifidobacteria which play a key role in the immune system89, has immunomodulatory51 and anti-inflammatory70,90 properties, and has an extensive and very positive safety profile91.
Study covers ivermectin, HCQ, and nitazoxanide.
1.
Gayozo et al., Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach, Biotecnia, doi:10.18633/biotecnia.v27.2485.
2.
Lefebvre et al., Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Bagheri-Far et al., Non-spike protein inhibition of SARS-CoV-2 by natural products through the key mediator protein ORF8, Molecular Biology Research Communications, doi:10.22099/mbrc.2024.50245.2001.
5.
de Oliveira Só et al., In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease, Preprints, doi:10.20944/preprints202404.1825.v1.
6.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
7.
Oranu et al., Validation of the binding affinities and stabilities of ivermectin and moxidectin against SARS-CoV-2 receptors using molecular docking and molecular dynamics simulation, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2024.26.1.0030.
8.
Zhao et al., Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis, Frontiers in Immunology, doi:10.3389/fimmu.2023.1197752.
9.
Vottero et al., Computational Prediction of the Interaction of Ivermectin with Fibrinogen, Molecular Sciences, doi:10.3390/ijms241411449.
10.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
11.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
12.
Alvarado et al., Interaction of the New Inhibitor Paxlovid (PF-07321332) and Ivermectin With the Monomer of the Main Protease SARS-CoV-2: A Volumetric Study Based on Molecular Dynamics, Elastic Networks, Classical Thermodynamics and SPT, Computational Biology and Chemistry, doi:10.1016/j.compbiolchem.2022.107692.
13.
Aminpour et al., In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds, Computation, doi:10.3390/computation10040051.
14.
Parvez et al., Insights from a computational analysis of the SARS-CoV-2 Omicron variant: Host–pathogen interaction, pathogenicity, and possible drug therapeutics, Immunity, Inflammation and Disease, doi:10.1002/iid3.639.
15.
Francés-Monerris et al., Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection, Physical Chemistry Chemical Physics, doi:10.1039/D1CP02967C.
16.
González-Paz et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry, doi:10.1016/j.bpc.2021.106677.
17.
González-Paz (B) et al., Structural Deformability Induced in Proteins of Potential Interest Associated with COVID-19 by binding of Homologues present in Ivermectin: Comparative Study Based in Elastic Networks Models, Journal of Molecular Liquids, doi:10.1016/j.molliq.2021.117284.
18.
Rana et al., A Computational Study of Ivermectin and Doxycycline Combination Drug Against SARS-CoV-2 Infection, Research Square, doi:10.21203/rs.3.rs-755838/v1.
19.
Muthusamy et al., Virtual Screening Reveals Potential Anti-Parasitic Drugs Inhibiting the Receptor Binding Domain of SARS-CoV-2 Spike protein, Journal of Virology & Antiviral Research, www.scitechnol.com/abstract/virtual-screening-reveals-potential-antiparasitic-drugs-inhibiting-the-receptor-binding-domain-of-sarscov2-spike-protein-16398.html.
20.
Qureshi et al., Mechanistic insights into the inhibitory activity of FDA approved ivermectin against SARS-CoV-2: old drug with new implications, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1906750.
21.
Schöning et al., Highly-transmissible Variants of SARS-CoV-2 May Be More Susceptible to Drug Therapy Than Wild Type Strains, Research Square, doi:10.21203/rs.3.rs-379291/v1.
22.
Bello et al., Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1911857.
23.
Udofia et al., In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV, Network Modeling Analysis in Health Informatics and Bioinformatics, doi:10.1007/s13721-021-00299-2.
24.
Choudhury et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Medicine, doi:10.2217/fvl-2020-0342.
25.
Kern et al., Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing, Frontiers in Pharmacology, doi:10.3389/fphar.2021.625678.
26.
Saha et al., The Binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2, Structural Chemistry, doi:10.1007/s11224-021-01776-0.
27.
Eweas et al., Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2, Frontiers in Microbiology, doi:10.3389/fmicb.2020.592908.
28.
Parvez (B) et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098.
29.
Francés-Monerris (B) et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
30.
Kalhor et al., Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816.
31.
Swargiary, A., Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies, Research Square, doi:10.21203/rs.3.rs-73308/v1.
32.
Maurya, D., A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients, American Chemical Society (ACS), doi:10.26434/chemrxiv.12630539.v1.
33.
Lehrer et al., Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, In Vivo, 34:5, 3023-3026, doi:10.21873/invivo.12134.
34.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
35.
Kofler et al., M-Motif, a potential non-conventional NLS in YAP/TAZ and other cellular and viral proteins that inhibits classic protein import, iScience, doi:10.1016/j.isci.2025.112105.
36.
Shahin et al., The selective effect of Ivermectin on different human coronaviruses; in-vitro study, Research Square, doi:10.21203/rs.3.rs-4180797/v1.
37.
Jitobaom et al., Identification of inositol monophosphatase as a broad‐spectrum antiviral target of ivermectin, Journal of Medical Virology, doi:10.1002/jmv.29552.
38.
Fauquet et al., Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction, Molecules, doi:10.3390/molecules28248072.
39.
García-Aguilar et al., In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction, International Journal of Molecular Sciences, doi:10.3390/ijms242216392.
40.
Liu et al., SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes, Stem Cell Research & Therapy, doi:10.1186/s13287-023-03485-3.
41.
Boschi et al., SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects, bioRxiv, doi:10.1101/2022.11.24.517882.
42.
De Forni et al., Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients, PLoS ONE, doi:10.1371/journal.pone.0276751.
43.
Saha (B) et al., Manipulation of Spray-Drying Conditions to Develop an Inhalable Ivermectin Dry Powder, Pharmaceutics, doi:10.3390/pharmaceutics14071432.
44.
Jitobaom (B) et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
45.
Croci et al., Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin, International Journal of Biomaterials, doi:10.1155/2016/8043983.
46.
Zheng et al., Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121719.
47.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
48.
Liu (B) et al., Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes, Stem Cell Reports, doi:10.1016/j.stemcr.2022.01.014.
49.
Segatori et al., Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients, Viruses, doi:10.3390/v13102084.
50.
Jitobaom (C) et al., Favipiravir and Ivermectin Showed in Vitro Synergistic Antiviral Activity against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-941811/v1.
51.
Munson et al., Niclosamide and ivermectin modulate caspase-1 activity and proinflammatory cytokine secretion in a monocytic cell line, British Society For Nanomedicine Early Career Researcher Summer Meeting, 2021, web.archive.org/web/20230401070026/https://michealmunson.github.io/COVID.pdf.
52.
Mountain Valley MD, Mountain Valley MD Receives Successful Results From BSL-4 COVID-19 Clearance Trial on Three Variants Tested With Ivectosol™, 5/18, www.globenewswire.com/en/news-release/2021/05/18/2231755/0/en/Mountain-Valley-MD-Receives-Successful-Results-From-BSL-4-COVID-19-Clearance-Trial-on-Three-Variants-Tested-With-Ivectosol.html.
53.
Yesilbag et al., Ivermectin also inhibits the replication of bovine respiratory viruses (BRSV, BPIV-3, BoHV-1, BCoV and BVDV) in vitro, Virus Research, doi:10.1016/j.virusres.2021.198384.
54.
Mody et al., Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents, Communications Biology, doi:10.1038/s42003-020-01577-x.
55.
Jeffreys et al., Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro activity against SARS-CoV-2, International Journal of Antimicrobial Agents, doi:10.1016/j.ijantimicag.2022.106542.
56.
Surnar et al., Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19, ACS Pharmacol. Transl. Sci., doi:10.1021/acsptsci.0c00179.
57.
Li et al., Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment, J. Cellular Physiology, doi:10.1002/jcp.30055.
58.
Caly et al., The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Research, doi:10.1016/j.antiviral.2020.104787.
59.
Zhang et al., Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice, Inflammation Research, doi:10.1007/s00011-008-8007-8.
60.
Gao et al., Ivermectin ameliorates acute myocarditis via the inhibition of importin-mediated nuclear translocation of NF-κB/p65, International Immunopharmacology, doi:10.1016/j.intimp.2024.112073.
61.
Abd-Elmawla et al., Suppression of NLRP3 inflammasome by ivermectin ameliorates bleomycin-induced pulmonary fibrosis, Journal of Zhejiang University-SCIENCE B, doi:10.1631/jzus.B2200385.
62.
Uematsu et al., Prophylactic administration of ivermectin attenuates SARS-CoV-2 induced disease in a Syrian Hamster Model, The Journal of Antibiotics, doi:10.1038/s41429-023-00623-0.
63.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
64.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
65.
Madrid et al., Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation, Heliyon, doi:10.1016/j.heliyon.2020.e05820.
66.
Ma et al., Ivermectin contributes to attenuating the severity of acute lung injury in mice, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113706.
67.
de Melo et al., Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin, EMBO Mol. Med., doi:10.15252/emmm.202114122.
68.
Arévalo et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Scientific Reports, doi:10.1038/s41598-021-86679-0.
69.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
70.
Yan et al., Anti-inflammatory effects of ivermectin in mouse model of allergic asthma, Inflammation Research, doi:10.1007/s00011-011-0307-8.
71.
Götz et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports, doi:10.1038/srep23138.
72.
Tay et al., Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Research, doi:10.1016/j.antiviral.2013.06.002.
73.
Wagstaff et al., Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal, doi:10.1042/BJ20120150.
74.
Wagstaff (B) et al., An AlphaScreen®-Based Assay for High-Throughput Screening for Specific Inhibitors of Nuclear Import, SLAS Discovery, doi:10.1177/1087057110390360.
75.
Barrows et al., A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection, Cell Host & Microbe, doi:10.1016/j.chom.2016.07.004.
76.
Yang et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research, doi:10.1016/j.antiviral.2020.104760.
77.
Mastrangelo et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, Journal of Antimicrobial Chemotherapy, doi:10.1093/jac/dks147.
78.
Varghese et al., Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses, Antiviral Research, doi:10.1016/j.antiviral.2015.12.012.
79.
Bennett et al., Role of a nuclear localization signal on the minor capsid Proteins VP2 and VP3 in BKPyV nuclear entry, Virology, doi:10.1016/j.virol.2014.10.013.
80.
Kosyna et al., The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways, Biological Chemistry, doi:10.1515/hsz-2015-0171.
81.
Scheim et al., Sialylated Glycan Bindings from SARS-CoV-2 Spike Protein to Blood and Endothelial Cells Govern the Severe Morbidities of COVID-19, International Journal of Molecular Sciences, doi:10.3390/ijms242317039.
82.
Liu (C) et al., Crosstalk between neutrophil extracellular traps and immune regulation: insights into pathobiology and therapeutic implications of transfusion-related acute lung injury, Frontiers in Immunology, doi:10.3389/fimmu.2023.1324021.
83.
Shouman et al., SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147, Cell Communication and Signaling, doi:10.1186/s12964-024-01718-3.
84.
Scheim (B), D., Ivermectin for COVID-19 Treatment: Clinical Response at Quasi-Threshold Doses Via Hypothesized Alleviation of CD147-Mediated Vascular Occlusion, SSRN, doi:10.2139/ssrn.3636557.
85.
Scheim (C), D., From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate and Then Clot Blood Cells, Center for Open Science, doi:10.31219/osf.io/sgdj2.
86.
Behl et al., CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target, Science of The Total Environment, doi:10.1016/j.scitotenv.2021.152072.
87.
DiNicolantonio et al., Ivermectin may be a clinically useful anti-inflammatory agent for late-stage COVID-19, Open Heart, doi:10.1136/openhrt-2020-001350.
88.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
89.
Hazan et al., Treatment with Ivermectin Increases the Population of Bifidobacterium in the Gut, ACG 2023, acg2023posters.eventscribe.net/posterspeakers.asp.
Arshad et al., 20 May 2020, peer-reviewed, 22 authors.
Prioritization of Anti‐SARS‐Cov‐2 Drug Repurposing Opportunities Based on Plasma and Target Site Concentrations Derived from their Established Human Pharmacokinetics
Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1909
There is a rapidly expanding literature on the in vitro antiviral activity of drugs that may be repurposed for therapy or chemoprophylaxis against severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2). However, this has not been accompanied by a comprehensive evaluation of the target plasma and lung concentrations of these drugs following approved dosing in humans. Accordingly, concentration 90% (EC 90 ) values recalculated from in vitro anti-SARS-CoV-2 activity data was expressed as a ratio to the achievable maximum plasma concentration (C max ) at an approved dose in humans (C max /EC 90 ratio). Only 14 of the 56 analyzed drugs achieved a C max /EC 90 ratio above 1. A more in-depth assessment demonstrated that only nitazoxanide, nelfinavir, tipranavir (ritonavir-boosted), and sulfadoxine achieved plasma concentrations above their reported anti-SARS-CoV-2 activity across their entire approved dosing interval. An unbound lung to plasma tissue partition coefficient (K p U lung ) was also simulated to derive a lung C max /half-maximal effective concentration (EC 50 ) as a better indicator of potential human efficacy. Hydroxychloroquine, chloroquine, mefloquine, atazanavir (ritonavirboosted), tipranavir (ritonavir-boosted), ivermectin, azithromycin, and lopinavir (ritonavir-boosted) were all predicted to achieve lung concentrations over 10-fold higher than their reported EC 50 . Nitazoxanide and sulfadoxine also exceeded their reported EC 50 by 7.8-fold and 1.5-fold in lung, respectively. This analysis may be used to select potential candidates for further clinical testing, while deprioritizing compounds unlikely to attain target concentrations for antiviral activity. Future studies should focus on EC 90 values and discuss findings in the context of achievable exposures in humans, especially within target compartments, such as the lungs, in order to maximize the potential for success of proposed human clinical trials.
SUPPORTING INFORMATION Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
ACKNOWLEDGMENTS The authors thank Nathan Morin from Alberta Health Services for being proactive in making them aware of previously published data for indomethacin. The authors also thank Articulate Science for publication support. CONFLICT OF INTEREST D.J.B. has received honoraria or advisory board payments from AbbVie, Gilead, ViiV, Merck, Janssen, and educational grants from AbbVie, Gilead, ViiV, Merck, Janssen, and Novartis. A.O. and S.P.R. are Directors of Tandem Nano Ltd. A.O. has received research funding from ViiV, Merck, Janssen, and consultancy from Gilead, ViiV and Merck not related to the current paper. P.O.N. is currently engaged in a collaboration with Romark LLC but this interaction did not influence the prioritization or conclusions in the current paper. All other authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS All authors wrote the paper. A.O. designed the research. U.A., H.B., L.T., H.P., and A.O. performed the research. R.R., H.P., U.A., and A.O. analyzed the data.
References
Amici, La Frazia, Brunelli, Balsamo, Angelini et al., Inhibition of viral protein translation by indomethacin in vesicular stomatitis virus infection: role of eIF2alpha kinase PKR, Cell Microbiol
Anderson, Curran, Nitazoxanide, None, Drugs
Ben-Zvi, Kivity, Langevitz, Shoenfeld, Hydroxychloroquine: from malaria to autoimmunity, Clin. Rev. Allergy Immunol
Bland, Altman, Statistical methods for assessing agreement between two methods of clinical measurement, Lancet
Bojkova, SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles, J bioRxiv, doi:10.1101/2020.04.03.024257
Browning, Pharmacology of chloroquine and hydroxychloroquine
Bukreyeva, Mantlo, Sattler, Huang, Paessler et al., The IMPDH inhibitor merimepodib suppresses SARS-CoV-2 replication in vitro, J bioRxiv, doi:10.1101/2020.04.07.028589
Burger, Agarwala, Child, Been-Tiktak, Wang et al., Effect of rifampin on steady-state pharmacokinetics of atazanavir with ritonavir in healthy volunteers, Antimicrobial Agents Chemother
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res
Chan-Tack, Struble, Birnkrant, Intracranial hemorrhage and liver-associated deaths associated with tipranavir/ritonavir: review of cases from the FDA's Adverse Event Reporting System, AIDS Patient Care STDS
Chandrasekaran, Ahmad, Shen, Demaio, Hultin et al., Disposition of bazedoxifene in rats, Xenobiotica
Chen, Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients, Clin. Infect. Dis, doi:10.1101/2020.02.29.20029520
Chen, Favipiravir versus Arbidol for COVID-19: a randomized clinical trial, J. medRxiv, doi:10.1101/2020.03.17.20037432.
Chen, Mook, Premont, Wang, Niclosamide: beyond an antihelminthic drug, Cell. Signal
Choy, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro, Antiviral. Res
Clerici, Trabattoni, Pacei, Biasin, Rossignol, The anti-infective nitazoxanide shows strong immumodulating effects, J. Immuno
Corbett, Lim, Kashuba, Kaletra (lopinavir/ ritonavir), Ann. Pharmacother
Damle, Stogniew, Dowell, Pharmacokinetics and tissue distribution of anidulafungin in rats, Antimicrob. Agents Chemother
De Kock, Pharmacokinetics of sulfadoxine and pyrimethamine for intermittent preventive treatment of malaria during pregnancy and after delivery, CPT Pharmacometrics Syst. Pharmacol
Diao, 2 (SARS-CoV-2) Infection, doi:10.1101/2020.03.04.20031120
Drachman, Kadlecek, Pourfathi, Xin, Profka et al., In vivo pH mapping of injured lungs using hyperpolarized [1-(13) C]pyruvate, Magn. Reson. Med
Fintelman-Rodrigues, Atazanavir inhibits SARS-CoV-2 replication and pro-inflammatory cytokine production, J bioRxiv, doi:10.1101/2020.04.04.020925
Fitch, Chevli, Gonzalez, Chloroquine-resistant Plasmodium falciparum: effect of substrate on chloroquine and amodiaquin accumulation, Antimicrob. Agents Chemother
Flexner, Bate, Kirkpatrick, Tipranavir, None, Nat. Rev. Drug Discovery
Fox, Saravolatz, Nitazoxanide: a new thiazolide antiparasitic agent, Clin. Infect. Dis
Garnock-Jones, Eltrombopag, None, Drugs
Gassen, SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection, Nat. Commun
Ge, A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19, J bioRxiv, doi:10.1101/2020.03.11.986836.
Geary, Divo, Jensen, Zangwill, Ginsburg, Kinetic modelling of the response of Plasmodium falciparum to chloroquine and its experimental testing in vitro. Implications for mechanism of action of and resistance to the drug, Biochem. Pharmacol
Gekonge, Bardin, Montaner, Short communication: Nitazoxanide inhibits HIV viral replication in monocyte-derived macrophages, AIDS Res. Hum. Retroviruses
Ginsburg, Nissani, Krugliak, Alkalinization of the food vacuole of malaria parasites by quinoline drugs and alkylamines is not correlated with their antimalarial activity, Biochem. Pharmacol
Goel, Gerriets, Chloroquine, StatPearls
Gonzalez, Schmidt, Derendorf, Importance of relating efficacy measures to unbound drug concentrations for antiinfective agents, Clin. Microbiol. Rev
Grein, Compassionate use of remdesivir for patients with severe Covid-19, N. Engl. J. Med, doi:10.1056/NEJMoa2007016
Gu, Han, Wang, COVID-19: Gastrointestinal manifestations and potential fecal-oral transmission, Gastroenterology
Guo, The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak -an update on the status, Mil Med Res
Gupta, Tulsankar, Bhatta, Misra, Pharmacokinetics, metabolism, and partial biodistribution of "pincer therapeutic" nitazoxanide in mice following pulmonary delivery of inhalable particles, Mol. Pharm
Haffizulla, Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial, Lancet Infect Dis
Harrison, Scott, Atazanavir, None, Drugs
Hickson, Margineantu, Hockenbery, Simon, Geballe, Inhibition of vaccinia virus replication by nitazoxanide, Virology
Hirani, Raucy, Lasker, Conversion of the HIV protease inhibitor nelfinavir to a bioactive metabolite by human liver CYP2C19, Drug Metab. Dispos
Hoffmann, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell
James, Nelfinavir (Viracept) approved: fourth protease inhibitor available, AIDS Treat. News
Jeon, Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs, J bioRxiv, doi:10.1101/2020.03.20.999730.
Jin, Structure of Mpro from COVID-19 virus and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Jurgeit, Mcdowell, Moese, Meldrum, Schwendener et al., Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects, PLoS Pathog
Justice, Drug toxicity, HIV progression, or comorbidity of aging: does tipranavir use increase the risk of intracranial hemorrhage?, Clin. Infect. Dis
Justice, Drug toxicity, HIV progression, or comorbidity of aging: does tipranavir use increase the risk of intracranial hemorrhage?, Clin. Infect. Dis
Kawai, Mathew, Tanaka, Rowland, Physiologically based pharmacokinetics of cyclosporine A: extension to tissue distribution kinetics in rats and scale-up to human, J. Pharmacol. Exp. Ther
King, Acosta, Tipranavir, None, Clin. Pharmacokinet
Krudsood, New fixed-dose artesunate-mefloquine formulation against multidrug-resistant Plasmodium falciparum in adults: a comparative phase iib safety and pharmacokinetic study with standard-dose nonfixed artesunate plus mefloquine, Antimicrob. Agents Chemother
Kruse, The steady-state pharmacokinetics of nelfinavir in combination with tenofovir in HIV-infected patients, Antivir. Ther
La Porte, Sabo, Beique, Cameron, Lack of effect of efavirenz on the pharmacokinetics of tipranavirritonavir in healthy volunteers, Antimicrob. Agents Chemother
Li, Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus, Nature
Li, Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer, PLoS One
Lien, Solheim, Ueland, Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment, Cancer Res
Lin, Preclinical evaluation of a nanoformulated antihelminthic, niclosamide, in ovarian cancer, Oncotarget
Liu, Ruhnke, Meersseman, Paiva, Kantecki et al., Pharmacokinetics of anidulafungin in critically ill patients with candidemia/invasive candidiasis, Antimicrob. Agents Chemother
Liu, Viral dynamics in mild and severe cases of COVID-19, Lancet Infect. Dis, doi:10.1016/S473-3099(20)30232-2
Lovegrove, Kain, Chapter 6 -malaria prevention
Lu, Liu, Jia, -nCoV transmission through the ocular surface must not be ignored, Lancet
Lv, Chu, Wang, HIV protease inhibitors: a review of molecular selectivity and toxicity, HIV AIDS
Markowitz, Long-term efficacy and safety of tipranavir boosted with ritonavir in HIV-1-infected patients failing multiple protease inhibitor regimens: 80-week data from a phase 2 study, J. Acquir. Immune Defic. Syndr
Mcchesney, Banks, Jr & Fabian, Tissue distribution of chloroquine, hydroxychloroquine, and desethylchloroquine in the rat, Toxicol. Appl. Pharmacol
Mchutchison, A randomized, double-blind, placebocontrolled dose-escalation trial of merimepodib (VX-497) and interferon-alpha in previously untreated patients with chronic hepatitis C, Antivir. Ther
Miller, Kok, Li, The virus inoculum volume influences outcome of influenza A infection in mice, Lab. Anim
Miller, Lobel, Satriale, Kuritsky, Stern et al., Severe cutaneous reactions among American travelers using pyrimethamine-sulfadoxine (Fansidar) for malaria prophylaxis, Am. J. Trop. Med. Hyg
Miner, Drug repurposing: the anthelmintics niclosamide and nitazoxanide are potent TMEM16A antagonists that fully bronchodilate airways, Front. Pharmacol
Moss, Wagner, Kanaoka, Olson, Yueh et al., The comparative disposition and metabolism of dolutegravir, a potent HIV-1 integrase inhibitor, in mice, rats, and monkeys, Xenobiotica
Motoya, Characterization of nelfinavir binding to plasma proteins and the lack of drug displacement interactions, HIV Med
Murdoch, Plosker, Anidulafungin, None, Drugs
Na-Bangchang, Limpaibul, Thanavibul, Tan-Ariya, Karbwang, The pharmacokinetics of chloroquine in healthy Thai subjects and patients with Plasmodium vivax malaria, Br. J. Clin. Pharmacol
Nalamachu, Wortmann, Role of indomethacin in acute pain and inflammation management: a review of the literature, Postgrad. Med
Orman, Perry, Tipranavir: a review of its use in the management of HIV infection, Drugs
Pober, Sessa, Evolving functions of endothelial cells in inflammation, Nat. Rev. Immunol
Porche, Ritonavir, Norvir), J. Assoc. Nurses AIDS Care
Price, Artesunate/mefloquine treatment of multi-drug resistant falciparum malaria, Trans. R Soc. Trop. Med. Hyg
Pugin, Dunn-Siegrist, Dufour, Tissieres, Charles et al., Cyclic stretch of human lung cells induces an acidification and promotes bacterial growth, Am. J. Respir. Cell Mol. Biol
Qin, Dysregulation of immune response in patients with COVID-19 in Wuhan, China, Clin. Infect. Dis, doi:10.1093/cid/ciaa248
Rainsford, Effects of misoprostol on the pharmacokinetics of indomethacin in human volunteers, Clin. Pharmacol. Ther
Raja, Lebbos, Kirkpatrick, Atazanavir sulphate, Nat. Rev. Drug Discov
Ritchie, Block, Nevin, Psychiatric side effects of mefloquine: applications to forensic psychiatry, J. Am. Acad. Psychiatry Law
Rivulgo, Comparative plasma exposure and lung distribution of two human use commercial azithromycin formulations assessed in murine model: a preclinical study, Biomed. Res. Int
Rodgers, Leahy, Rowland, Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases, J. Pharm. Sci
Rodgers, Rowland, Mechanistic approaches to volume of distribution predictions: understanding the processes, Pharm. Res
Rodgers, Rowland, Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions, J. Pharm. Sci
Rossignol, La Frazia, Chiappa, Ciucci, Santoro et al., a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level, J. Biol. Chem
Rossignol, Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus, J. Infect. Public Health
Rossignol, Nitazoxanide: a first-in-class broad-spectrum antiviral agent, Antiviral Res
Sanders, Monogue, Jodlowski, Cutrell, pharmacologic treatments for coronavirus disease, doi:10.1001/jama.2020.6019
Shida, Takahashi, Nohda, Hirama, Pharmacokinetics and pharmacodynamics of eltrombopag in healthy Japanese males, Jpn. J. Clin. Pharmacol. Ther
Smith, Kulkarni, Chen, Goldstein, Influenza virus inoculum volume is critical to elucidate age-dependent mortality in mice, Aging Cell
Streeck, Rockstroh, Review of tipranavir in the treatment of drug-resistant HIV, Ther. Clin. Risk Manag
Sun, Wang, Liu, Liu, Role of the Eye in Transmitting Human Coronavirus: What We Know and What We Do Not Know, Frontiers in Public Health, doi:10.3389/fpubh.2020.00155
Tett, Cutler, Beck, Day, Concentration-effect relationship of hydroxychloroquine in patients with rheumatoid arthritis-a prospective, dose ranging study, J. Rheumatol
Tilmanis, Van Baalen, Oh, Rossignol, Hurt, The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide, Antiviral Res
Touret, In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication, J bioRxiv, doi:10.1101/2020.04.03.023846
Trabattoni, Thiazolides elicit anti-viral innate immunity and reduce HIV replication, Sci. Rep
Unis, Mitochondrial mechanisms of nelfinavir toxicity in human brain microvascular endothelial cells, The FASEB Journal
Vanderkooi, Prapunwattana, Yuthavong, Evidence for electrogenic accumulation of mefloquine by malarial parasites, Biochem. Pharmacol
Wang, Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission, Antiviral Res
Wang, Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res
Weston, Haupt, Logue, Matthews, Frieman, FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro, J bioRxiv, doi:10.1101/2020.03.25.008482
Wong, Lui, Sung, COVID-19 and the digestive system, J. Gastroenterol. Hepatol
Wu, Prolonged presence of SARS-CoV-2 viral RNA in faecal samples, Lancet Gastroenterol. Hepatol, doi:10.1016/S2468-1253(20)30083-2.
Xu, Gao, Wu, Selinger, Zhou, Indomethacin has a potent antiviral activity against SARS CoV-2 in vitro and canine coronavirus in vivo, J bioRxiv, doi:10.1101/2020.04.01.017624
Xu, Nelfinavir is active against SARS-CoV-2 in Vero E6, Cells, doi:.10.26434/chemrxiv.12039888
Yamamoto, Matsuyama, Hoshino, Yamamoto, Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro, J bioRxiv, doi:10.1101/2020.04.06.026476
Yao, In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis, doi:10.1093/cid/ciaa237
Yeh, Pharmacokinetic overview of indomethacin and sustained-release indomethacin, Am. J. Med
Zhang, Circulating metabolites of the human immunodeficiency virus protease inhibitor nelfinavir in humans: structural identification, levels in plasma, and antiviral activities, Antimicrob. Agents Chemother
Zhang, Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes, Emerg. Microbes Infect
Zhang, Shi, Wang, Liver injury in COVID-19: management and challenges, Lancet Gastroenterol. Hepatol
Zhang, Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins using highresolution mass spectrometry, J bioRxiv, doi:10.1101/2020.03.28.013276.
Zhang, The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes, J bioRxiv, doi:10.1101/2020.01.30.927806.
Zhang, Tumor-associated inflammatory microenvironment in non-small cell lung cancer: correlation with FGFR1 and TLR4 expression via PI3K/Akt pathway, J. Cancer
Zhang, Yap, Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs, Bioorg. Med. Chem
Zhao, Zhao, Wang, Zhou, Ma et al., Singlecell RNA expression profiling of ACE2, the receptor of SARS-CoV-2, J bioRxiv
DOI record:
{
"DOI": "10.1002/cpt.1909",
"ISSN": [
"0009-9236",
"1532-6535"
],
"URL": "http://dx.doi.org/10.1002/cpt.1909",
"alternative-id": [
"10.1002/cpt.1909"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2020-04-24"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2020-05-18"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2020-06-14"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-1586-1885",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Arshad",
"given": "Usman",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Pertinez",
"given": "Henry",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Box",
"given": "Helen",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Tatham",
"given": "Lee",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Rajoli",
"given": "Rajith K. R.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4596-2708",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Curley",
"given": "Paul",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4960-2139",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Neary",
"given": "Megan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Sharp",
"given": "Joanne",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5980-8966",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Liptrott",
"given": "Neill J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Valentijn",
"given": "Anthony",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8504-2354",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "David",
"given": "Christopher",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6946-1097",
"affiliation": [
{
"name": "Department of Chemistry University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Rannard",
"given": "Steve P.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Chemistry University of Liverpool Liverpool UK"
}
],
"family": "O’Neill",
"given": "Paul M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5288-4501",
"affiliation": [
{
"name": "Department of Tropical Disease Biology Liverpool School of Tropical Medicine Centre for Drugs and Diagnostics Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Aljayyoussi",
"given": "Ghaith",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7160-6275",
"affiliation": [
{
"name": "Department of Tropical Disease Biology Liverpool School of Tropical Medicine Centre for Drugs and Diagnostics Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Pennington",
"given": "Shaun H.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2331-3192",
"affiliation": [
{
"name": "Department of Tropical Disease Biology Liverpool School of Tropical Medicine Centre for Drugs and Diagnostics Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Ward",
"given": "Stephen A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Hill",
"given": "Andrew",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Back",
"given": "David J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"family": "Khoo",
"given": "Saye H.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pat Bray Electrical Wigan UK"
}
],
"family": "Bray",
"given": "Patrick G.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6356-6595",
"affiliation": [
{
"name": "Department of Tropical Disease Biology Liverpool School of Tropical Medicine Centre for Drugs and Diagnostics Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Biagini",
"given": "Giancarlo A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9819-7651",
"affiliation": [
{
"name": "Department of Molecular and Clinical Pharmacology Materials Innovation Factory University of Liverpool Liverpool UK"
}
],
"authenticated-orcid": false,
"family": "Owen",
"given": "Andrew",
"sequence": "additional"
}
],
"container-title": "Clinical Pharmacology & Therapeutics",
"container-title-short": "Clin. Pharmacol. Ther.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2020,
5,
21
]
],
"date-time": "2020-05-21T22:51:38Z",
"timestamp": 1590101498000
},
"deposited": {
"date-parts": [
[
2020,
9,
19
]
],
"date-time": "2020-09-19T11:44:50Z",
"timestamp": 1600515890000
},
"indexed": {
"date-parts": [
[
2022,
8,
16
]
],
"date-time": "2022-08-16T16:26:41Z",
"timestamp": 1660667201410
},
"is-referenced-by-count": 84,
"issue": "4",
"issued": {
"date-parts": [
[
2020,
6,
14
]
]
},
"journal-issue": {
"issue": "4",
"published-print": {
"date-parts": [
[
2020,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
6,
14
]
],
"date-time": "2020-06-14T00:00:00Z",
"timestamp": 1592092800000
}
},
{
"URL": "http://doi.wiley.com/10.1002/tdm_license_1.1",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
6,
14
]
],
"date-time": "2020-06-14T00:00:00Z",
"timestamp": 1592092800000
}
}
],
"link": [
{
"URL": "https://api.wiley.com/onlinelibrary/tdm/v1/articles/10.1002%2Fcpt.1909",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1002/cpt.1909",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/full-xml/10.1002/cpt.1909",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1002/cpt.1909",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "311",
"original-title": [],
"page": "775-790",
"prefix": "10.1002",
"published": {
"date-parts": [
[
2020,
6,
14
]
]
},
"published-online": {
"date-parts": [
[
2020,
6,
14
]
]
},
"published-print": {
"date-parts": [
[
2020,
10
]
]
},
"publisher": "Wiley",
"reference": [
{
"DOI": "10.1053/j.gastro.2020.02.054",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_1_1"
},
{
"key": "e_1_2_11_2_1",
"unstructured": "World Health Organisation.COVID‐19 Trials ‐ International Clinical Trials Registry Platform (ICTRP)<https://www.who.int/ictrp/search/en/> (2020)."
},
{
"article-title": "Dysregulation of immune response in patients with COVID‐19 in Wuhan, China",
"author": "Qin C.",
"journal-title": "Clin. Infect. Dis.",
"key": "e_1_2_11_3_1"
},
{
"DOI": "10.1038/nature02145",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_4_1"
},
{
"article-title": "Single‐cell RNA expression profiling of ACE2, the receptor of SARS‐CoV‐2",
"author": "Zhao Y.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_5_1"
},
{
"article-title": "Prolonged presence of SARS‐CoV‐2 viral RNA in faecal samples",
"author": "Wu Y.",
"journal-title": "Lancet Gastroenterol. Hepatol.",
"key": "e_1_2_11_6_1"
},
{
"article-title": "Kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) Infection",
"author": "Diao B.H.",
"journal-title": "J medRxiv",
"key": "e_1_2_11_7_1"
},
{
"DOI": "10.1016/S2468-1253(20)30057-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_8_1"
},
{
"DOI": "10.1186/s40779-020-00240-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_9_1"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_10_1"
},
{
"article-title": "The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes",
"author": "Zhang H.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_11_1"
},
{
"DOI": "10.1111/jgh.15047",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_12_1"
},
{
"DOI": "10.1080/22221751.2020.1729071",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_13_1"
},
{
"DOI": "10.3389/fpubh.2020.00155",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_14_1"
},
{
"DOI": "10.1016/S0140-6736(20)30313-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_15_1"
},
{
"DOI": "10.1111/acel.12893",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_16_1"
},
{
"DOI": "10.1258/la.2012.011157",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_17_1"
},
{
"DOI": "10.1101/2020.02.29.20029520",
"doi-asserted-by": "crossref",
"key": "e_1_2_11_18_1",
"unstructured": "Chen X.et al.Detectable serum SARS‐CoV‐2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL‐6) level in critically ill COVID‐19 patients.Clin. Infect. Dis.https://doi.org/10.1101/2020.02.29.20029520."
},
{
"article-title": "Viral dynamics in mild and severe cases of COVID‐19",
"author": "Liu Y.",
"journal-title": "Lancet Infect. Dis.",
"key": "e_1_2_11_19_1"
},
{
"DOI": "10.1002/jps.20322",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_20_1"
},
{
"DOI": "10.1002/jps.20502",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_21_1"
},
{
"DOI": "10.1007/s11095-006-9210-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_22_1"
},
{
"DOI": "10.1128/AAC.01596-07",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_23_1"
},
{
"DOI": "10.3109/00498254.2010.492879",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_24_1"
},
{
"DOI": "10.1007/978-1-4939-0597-3_2",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_25_1"
},
{
"DOI": "10.1016/0041-008X(67)90089-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_26_1"
},
{
"DOI": "10.1021/acs.molpharmaceut.6b01089",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_27_1"
},
{
"article-title": "Distribution of tamoxifen and its metabolites in rat and human tissues during steady‐state treatment",
"author": "Lien E.A.",
"first-page": "4837",
"journal-title": "Cancer Res.",
"key": "e_1_2_11_28_1",
"volume": "51",
"year": "1991"
},
{
"DOI": "10.1155/2013/392010",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_29_1"
},
{
"DOI": "10.3109/00498254.2014.942409",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_30_1"
},
{
"article-title": "Physiologically based pharmacokinetics of cyclosporine A: extension to tissue distribution kinetics in rats and scale‐up to human",
"author": "Kawai R.",
"first-page": "457",
"journal-title": "J. Pharmacol. Exp. Ther.",
"key": "e_1_2_11_31_1",
"volume": "287",
"year": "1998"
},
{
"key": "e_1_2_11_32_1",
"unstructured": "Pharmaceuticals and Medical Devices Agency(PMDA).Report on the deliberation results ‐ Xospata tablets 40 mg<https://www.pmda.go.jp/drugs/2014/P201400148/800155000_22600AMX01325_I100_1.pdf> (2018). Accessed May 4 2020."
},
{
"DOI": "10.1016/S0140-6736(86)90837-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_33_1"
},
{
"article-title": "FDA approved drugs with broad anti‐coronaviral activity inhibit SARS‐CoV‐2 in vitro",
"author": "Weston S.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_34_1"
},
{
"article-title": "A data‐driven drug repositioning framework discovered a potential therapeutic agent targeting COVID‐19",
"author": "Ge Y.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_35_1"
},
{
"article-title": "SARS‐CoV‐2 and SARS‐CoV differ in their cell tropism and drug sensitivity profiles",
"author": "Bojkova D.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_36_1"
},
{
"article-title": "In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS‐CoV‐2 replication",
"author": "Touret F.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_37_1"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_38_1"
},
{
"article-title": "Identification of antiviral drug candidates against SARS‐CoV‐2 from FDA‐approved drugs",
"author": "Jeon S.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_39_1"
},
{
"article-title": "Indomethacin has a potent antiviral activity against SARS CoV‐2 in vitro and canine coronavirus in vivo",
"author": "Xu T.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_40_1"
},
{
"article-title": "Atazanavir inhibits SARS‐CoV‐2 replication and pro‐inflammatory cytokine production",
"author": "Fintelman‐Rodrigues N.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_41_1"
},
{
"article-title": "Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro",
"author": "Yamamoto N.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_42_1"
},
{
"article-title": "The IMPDH inhibitor merimepodib suppresses SARS‐CoV‐2 replication in vitro",
"author": "Bukreyeva N.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_43_1"
},
{
"article-title": "Structure of Mpro from COVID‐19 virus and discovery of its inhibitors",
"author": "Jin Z.",
"journal-title": "Nature",
"key": "e_1_2_11_44_1"
},
{
"article-title": "In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)",
"author": "Yao X.",
"journal-title": "Clin. Infect. Dis.",
"key": "e_1_2_11_45_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104786",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_46_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_47_1"
},
{
"article-title": "Nelfinavir is active against SARS‐CoV‐2 in Vero E6",
"author": "Xu Z.",
"journal-title": "Cells",
"key": "e_1_2_11_48_1"
},
{
"DOI": "10.1101/2020.03.17.20037432",
"doi-asserted-by": "crossref",
"key": "e_1_2_11_49_1",
"unstructured": "Chen C.et al.Favipiravir versus Arbidol for COVID‐19: a randomized clinical trial.J. medRxiv.https://doi.org/10.1101/2020.03.17.20037432."
},
{
"article-title": "Compassionate use of remdesivir for patients with severe Covid‐19",
"author": "Grein J.",
"journal-title": "N. Engl. J. Med.",
"key": "e_1_2_11_50_1"
},
{
"DOI": "10.1016/j.jiph.2016.04.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_51_1"
},
{
"DOI": "10.1016/j.antiviral.2017.10.002",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_52_1"
},
{
"DOI": "10.1016/S1473-3099(14)70717-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_53_1"
},
{
"DOI": "10.1089/aid.2014.0015",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_54_1"
},
{
"DOI": "10.1038/srep27148",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_55_1"
},
{
"DOI": "10.1016/j.antiviral.2014.07.014",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_56_1"
},
{
"DOI": "10.1074/jbc.M109.029470",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_57_1"
},
{
"DOI": "10.1016/j.virol.2018.03.023",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_58_1"
},
{
"DOI": "10.1016/j.antiviral.2016.10.003",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_59_1"
},
{
"article-title": "Site‐specific N‐glycosylation characterization of recombinant SARS‐CoV‐2 spike proteins using high‐resolution mass spectrometry",
"author": "Zhang Y.",
"journal-title": "J bioRxiv",
"key": "e_1_2_11_60_1"
},
{
"key": "e_1_2_11_61_1",
"unstructured": "Clerici M. Trabattoni D. Pacei M. Biasin M.&Rossignol J.‐F.The anti‐infective nitazoxanide shows strong immumodulating effects. J. Immuno.186 155.21(2011)."
},
{
"DOI": "10.1371/journal.ppat.1002976",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_62_1"
},
{
"DOI": "10.1016/j.bmc.2004.03.035",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_63_1"
},
{
"DOI": "10.1038/s41467-019-13659-4",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_64_1"
},
{
"DOI": "10.1371/journal.pone.0074670",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_65_1"
},
{
"DOI": "10.18632/oncotarget.7113",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_66_1"
},
{
"DOI": "10.3389/fphar.2019.00051",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_67_1"
},
{
"article-title": "HIV protease inhibitors: a review of molecular selectivity and toxicity",
"author": "Lv Z.",
"first-page": "95",
"journal-title": "HIV AIDS (Auckl)",
"key": "e_1_2_11_68_1",
"volume": "7",
"year": "2015"
},
{
"article-title": "Review of tipranavir in the treatment of drug‐resistant HIV",
"author": "Streeck H.",
"first-page": "641",
"journal-title": "Ther. Clin. Risk Manag.",
"key": "e_1_2_11_69_1",
"volume": "3",
"year": "2007"
},
{
"article-title": "pharmacologic treatments for coronavirus disease 2019 (COVID‐19)",
"author": "Sanders J.M.",
"journal-title": "JAMA",
"key": "e_1_2_11_70_1"
},
{
"DOI": "10.1086/592302",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_71_1"
},
{
"DOI": "10.1038/nrd1907",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_72_1"
},
{
"DOI": "10.1089/apc.2008.0043",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_73_1"
},
{
"DOI": "10.1124/dmd.104.001743",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_74_1"
},
{
"DOI": "10.1128/AAC.45.4.1086-1093.2001",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_75_1"
},
{
"DOI": "10.1111/j.1468-1293.2006.00356.x",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_76_1"
},
{
"DOI": "10.2165/00003088-200645070-00003",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_77_1"
},
{
"DOI": "10.1097/QAI.0b013e318074eff5",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_78_1"
},
{
"DOI": "10.1086/592302",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_79_1"
},
{
"article-title": "Mitochondrial mechanisms of nelfinavir toxicity in human brain microvascular endothelial cells",
"author": "Unis G.",
"journal-title": "The FASEB Journal",
"key": "e_1_2_11_80_1",
"volume": "30",
"year": "2016"
},
{
"DOI": "10.1016/B978-141602613-6.10006-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_81_1"
},
{
"DOI": "10.1002/psp4.12181",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_82_1"
},
{
"DOI": "10.1111/cmi.12446",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_83_1"
},
{
"DOI": "10.3810/pgm.2014.07.2787",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_84_1"
},
{
"DOI": "10.1038/nri2171",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_85_1"
},
{
"DOI": "10.7150/jca.26277",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_86_1"
},
{
"DOI": "10.1016/0006-2952(90)90302-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_87_1"
},
{
"DOI": "10.1165/rcmb.2007-0114OC",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_88_1"
},
{
"DOI": "10.1002/mrm.26473",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_89_1"
},
{
"DOI": "10.1016/0006-2952(89)90550-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_90_1"
},
{
"DOI": "10.1016/0006-2952(88)90394-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_91_1"
},
{
"DOI": "10.1128/AAC.6.6.757",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_92_1"
},
{
"article-title": "Psychiatric side effects of mefloquine: applications to forensic psychiatry",
"author": "Ritchie E.C.",
"first-page": "224",
"journal-title": "J. Am. Acad. Psychiatry Law",
"key": "e_1_2_11_93_1",
"volume": "41",
"year": "2013"
},
{
"DOI": "10.1128/CMR.00092-12",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_94_1"
},
{
"DOI": "10.1086/428839",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_95_1"
},
{
"DOI": "10.1128/AAC.00462-09",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_96_1"
},
{
"article-title": "The steady‐state pharmacokinetics of nelfinavir in combination with tenofovir in HIV‐infected patients",
"author": "Kruse G.",
"first-page": "349",
"journal-title": "Antivir. Ther.",
"key": "e_1_2_11_97_1",
"volume": "10",
"year": "2005"
},
{
"DOI": "10.1038/clpt.1992.41",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_98_1"
},
{
"DOI": "10.1128/AAC.00461-06",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_99_1"
},
{
"article-title": "Concentration‐effect relationship of hydroxychloroquine in patients with rheumatoid arthritis–a prospective, dose ranging study",
"author": "Tett S.E.",
"first-page": "1656",
"journal-title": "J. Rheumatol.",
"key": "e_1_2_11_100_1",
"volume": "27",
"year": "2000"
},
{
"DOI": "10.3999/jscpt.42.11",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_101_1"
},
{
"author": "US Food and Drug Administration (FDA)",
"key": "e_1_2_11_102_1",
"volume-title": "Abbott L. Clinical Pharmacology and Biopharmaceutics review of Kaletra oral solution (NDA#021251)",
"year": "2020"
},
{
"DOI": "10.1111/j.1365-2125.1994.tb04354.x",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_103_1"
},
{
"DOI": "10.1128/AAC.01187-09",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_104_1"
},
{
"DOI": "10.1128/AAC.02139-12",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_105_1"
},
{
"DOI": "10.1038/nrd1232",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_106_1"
},
{
"DOI": "10.2165/00003495-200565160-00010",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_107_1"
},
{
"DOI": "10.2165/00003495-200464190-00011",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_108_1"
},
{
"key": "e_1_2_11_109_1",
"unstructured": "Goel P.&Gerriets V.Chloroquine. In:StatPearls. (StatPearls Publishing Treasure Island (FL).https://www.ncbi.nlm.nih.gov/books/NBK551512/2020."
},
{
"DOI": "10.2165/11207390-000000000-00000",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_110_1"
},
{
"author": "(PMDA)",
"key": "e_1_2_11_111_1",
"volume-title": "Report on the Deliberation Results."
},
{
"DOI": "10.1007/s12016-010-8243-x",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_112_1"
},
{
"DOI": "10.1016/0002-9343(85)90510-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_113_1"
},
{
"DOI": "10.1345/aph.1A363",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_114_1"
},
{
"DOI": "10.1016/S0035-9203(97)90032-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_115_1"
},
{
"article-title": "A randomized, double‐blind, placebo‐controlled dose‐escalation trial of merimepodib (VX‐497) and interferon‐alpha in previously untreated patients with chronic hepatitis C",
"author": "McHutchison J.G.",
"first-page": "635",
"journal-title": "Antivir. Ther.",
"key": "e_1_2_11_116_1",
"volume": "10",
"year": "2005"
},
{
"article-title": "Nelfinavir (Viracept) approved: fourth protease inhibitor available",
"author": "James J.S.",
"first-page": "1",
"journal-title": "AIDS Treat. News",
"key": "e_1_2_11_117_1",
"volume": "267",
"year": "1997"
},
{
"DOI": "10.1016/j.cellsig.2017.04.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_118_1"
},
{
"DOI": "10.2165/00003495-200767130-00015",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_119_1"
},
{
"DOI": "10.1016/S1055-3290(97)80061-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_120_1"
},
{
"DOI": "10.4269/ajtmh.1986.35.451",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_121_1"
},
{
"DOI": "10.2165/00003495-200868100-00006",
"doi-asserted-by": "publisher",
"key": "e_1_2_11_122_1"
}
],
"reference-count": 122,
"references-count": 122,
"relation": {},
"resource": {
"primary": {
"URL": "https://onlinelibrary.wiley.com/doi/10.1002/cpt.1909"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Pharmacology (medical)",
"Pharmacology"
],
"subtitle": [],
"title": "Prioritization of Anti‐SARS‐Cov‐2 Drug Repurposing Opportunities Based on Plasma and Target Site Concentrations Derived from their Established Human Pharmacokinetics",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy",
"volume": "108"
}