Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach
et al., Research Square, doi:10.21203/rs.3.rs-3888947/v1, Jan 2024
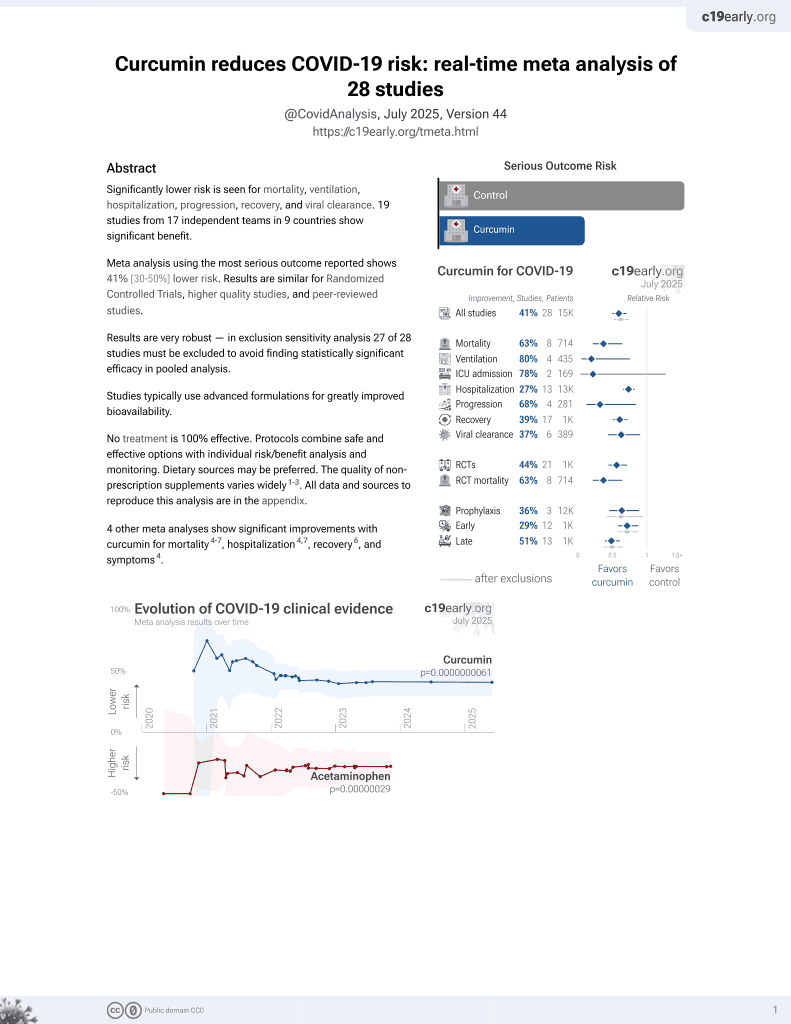
Curcumin for COVID-19
17th treatment shown to reduce risk in
February 2021, now with p = 0.0000000061 from 28 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico and in vitro study including quercetin and curcumin derivatives as potential SARS-CoV-2 main protease (Mpro) inhibitors. Molecular dynamics simulations and virtual screening identified quercetin and curcumin derivatives demethoxycurcumin and hexahydrocurcumin as potential binders of Mpro. Demethoxycurcumin was tested in vitro, showing significant inhibitory activity against SARS-CoV-2, with no cytotoxicity observed.
62 preclinical studies support the efficacy of curcumin for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with curcumin or metabolites via binding to the spikeA,1,5,6,11,16,18,24,27 (and specifically the receptor binding domainB,2,4,14,17,20 ), MproC,4-6,11,13,15-17,19,20,22,25,27,28,30,48 , RNA-dependent RNA polymeraseD,4-6,17,26 , PLproE,6, ACE2F,2,18,19,21 , nucleocapsidG,12,29 , nsp10H,29, and helicaseI,36 proteins, and inhibition of spike-ACE2 interactionJ,3.
In vitro studies demonstrate inhibition of the spikeA,41 (and specifically the receptor binding domainB,51), MproC,23,41,48,50 , ACE2F,51, and TMPRSS2K,51 proteins, and inhibition of spike-ACE2 interactionJ,3,34 .
In vitro studies demonstrate efficacy in Calu-3L,49, A549M,41, A549-ATN,31, 293TO,7, HEK293-hACE2P,23,39 , 293T/hACE2/TMPRSS2Q,40, Vero E6R,1,13,17,27,39,41,43,45,47,49 , and SH-SY5YS,38 cells.
Curcumin decreases pro-inflammatory cytokines induced by SARS-CoV-2 in peripheral blood mononuclear cells47, alleviates SARS-CoV-2 spike protein-induced mitochondrial membrane damage and oxidative stress7, may limit COVID-19 induced cardiac damage by inhibiting the NF-κB signaling pathway which mediates the profibrotic effects of the SARS-CoV-2 spike protein on cardiac fibroblasts35, is predicted to inhibit the interaction between the SARS-CoV-2 spike protein receptor binding domain and the human ACE2 receptor for the delta and omicron variants14, lowers ACE2 and STAT3, curbing lung inflammation and ARDS in preclinical COVID-19 models32, inhibits SARS-CoV-2 ORF3a ion channel activity, which contributes to viral pathogenicity and cytotoxicity42, has direct virucidal action by disrupting viral envelope integrity44, may inhibit viral replication and modulate inflammatory pathways like NF-κB via SIRT1 activation52, and can function as a photosensitizer in photodynamic therapy to generate reactive oxygen species that damage the virus44.
Study covers curcumin and quercetin.
1.
Marzouk et al., Computational and Experimental Insights into the Antiviral Mechanism of Turmeric (Curcuma longa) against SARS-CoV-2 D614G, BIO Web of Conferences, doi:10.1051/bioconf/202519804002.
2.
Wu et al., Utilizing natural compounds as ligands to disrupt the binding of SARS-CoV-2 receptor-binding domain to angiotensin-converting enzyme 2, impeding viral infection, Phytochemistry Letters, doi:10.1016/j.phytol.2025.102999.
3.
Najimi et al., Phytochemical Inhibitors of SARS‐CoV‐2 Entry: Targeting the ACE2‐RBD Interaction with l‐Tartaric Acid, l‐Ascorbic Acid, and Curcuma longa Extract, ChemistrySelect, doi:10.1002/slct.202406035.
4.
Rajamanickam et al., Exploring the Potential of Siddha Formulation MilagaiKudineer-Derived Phytotherapeutics Against SARS-CoV-2: An In-Silico Investigation for Antiviral Intervention, Journal of Pharmacy and Pharmacology Research, doi:10.26502/fjppr.0105.
5.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
6.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
7.
Zhang et al., Computational Discovery of Mitochondrial Dysfunction Biomarkers in Severe SARS-CoV-2 Infection: Facilitating Pytomedicine Screening, Phytomedicine, doi:10.1016/j.phymed.2024.155784.
8.
Öztürkkan et al., In Silico investigation of the effects of curcuminoids on the spike protein of the omicron variant of SARS-CoV-2, Baku State University Journal of Chemistry and Material Sciences, 1:2, bsuj.bsu.edu.az/uploads/pdf/ec4204d62f7802de54e6092bf7860029.pdf.
9.
Yunze et al., Therapeutic effect and potential mechanism of curcumin, an active ingredient in Tongnao Decoction, on COVID-19 combined with stroke: a network pharmacology study and GEO database mining, Research Square, doi:10.21203/rs.3.rs-4329762/v1.
10.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
11.
Boseila et al., Throat spray formulated with virucidal Pharmaceutical excipients as an effective early prophylactic or treatment strategy against pharyngitis post-exposure to SARS CoV-2, European Journal of Pharmaceutics and Biopharmaceutics, doi:10.1016/j.ejpb.2024.114279.
12.
Hidayah et al., Bioinformatics study of curcumin, demethoxycurcumin, bisdemethoxycurcumin and cyclocurcumin compounds in Curcuma longa as an antiviral agent via nucleocapsid on SARS-CoV-2 inhibition, International Conference on Organic and Applied Chemistry, doi:10.1063/5.0197724.
13.
Singh et al., Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach, Research Square, doi:10.21203/rs.3.rs-3888947/v1.
14.
Kant et al., Structure-based drug discovery to identify SARS-CoV2 spike protein–ACE2 interaction inhibitors, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2300060.
15.
Naderi Beni et al., In silico studies of anti-oxidative and hot temperament-based phytochemicals as natural inhibitors of SARS-CoV-2 Mpro, PLOS ONE, doi:10.1371/journal.pone.0295014.
16.
Moschovou et al., Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms242115894.
17.
Eleraky et al., Curcumin Transferosome-Loaded Thermosensitive Intranasal in situ Gel as Prospective Antiviral Therapy for SARS-Cov-2, International Journal of Nanomedicine, doi:10.2147/IJN.S423251.
18.
Singh (B) et al., Computational studies to analyze effect of curcumin inhibition on coronavirus D614G mutated spike protein, The Seybold Report, doi:10.17605/OSF.IO/TKEXJ.
19.
Thapa et al., In-silico Approach for Predicting the Inhibitory Effect of Home Remedies on Severe Acute Respiratory Syndrome Coronavirus-2, Makara Journal of Science, doi:10.7454/mss.v27i3.1609.
20.
Srivastava et al., Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach, Stresses, doi:10.3390/stresses3030043.
21.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
22.
Winih Kinasih et al., Analisis in silico interaksi senyawa kurkuminoid terhadap enzim main protease 6LU7 dari SARS-CoV-2, Duta Pharma Journal, doi:10.47701/djp.v3i1.2904.
23.
Wu (B) et al., Potential Mechanism of Curcumin and Resveratrol against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-2780614/v1.
24.
Nag et al., Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2022.105552.
25.
Rampogu et al., Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2, International Journal of Molecular Sciences, doi:10.3390/ijms23031771.
26.
Singh (C) et al., Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2021.104965.
27.
Kandeil et al., Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2, Pathogens, doi:10.3390/pathogens10060758.
28.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
29.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
30.
Sekiou et al., In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2, ChemRxiv, doi:10.26434/chemrxiv.12181404.v1.
31.
Grüneberg et al., Dose-dependent antiviral effects of glycyrrhizin, curcumin, and harmaline against clinical SARS-CoV-2 isolates, including D614G, Omicron BA.5, and Omicron XBB.1, BMC Complementary Medicine and Therapies, doi:10.1186/s12906-026-05253-1.
32.
Aktay et al., Oral Administration of Water-Soluble Curcumin Complex Prevents ARDS With the Potential for COVID-19 Treatment, Phytotherapy Research, doi:10.1002/ptr.70046.
33.
Olubiyi et al., Novel dietary herbal preparations with inhibitory activities against multiple SARS-CoV-2 targets: A multidisciplinary investigation into antiviral activities, Food Chemistry Advances, doi:10.1016/j.focha.2025.100969.
34.
Emam et al., Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors, AMB Express, doi:10.1186/s13568-024-01739-8.
35.
Van Tin et al., Spike Protein of SARS-CoV-2 Activates Cardiac Fibrogenesis through NLRP3 Inflammasomes and NF-κB Signaling, Cells, doi:10.3390/cells13161331.
36.
Li et al., Thermal shift assay (TSA)-based drug screening strategy for rapid discovery of inhibitors against the Nsp13 helicase of SARS-CoV-2, Animals and Zoonoses, doi:10.1016/j.azn.2024.06.001.
37.
Kamble et al., Nanoparticulate curcumin spray imparts prophylactic and therapeutic properties against SARS-CoV-2, Emergent Materials, doi:10.1007/s42247-024-00754-6.
38.
Nicoliche et al., Antiviral, anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-61662-7.
39.
Nittayananta et al., A novel film spray containing curcumin inhibits SARS-CoV-2 and influenza virus infection and enhances mucosal immunity, Virology Journal, doi:10.1186/s12985-023-02282-x.
40.
Septisetyani et al., Curcumin and turmeric extract inhibited SARS-CoV-2 pseudovirus cell entry and Spike mediated cell fusion, bioRxiv, doi:10.1101/2023.09.28.560070.
41.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
42.
Fam et al., Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Scientific Reports, doi:10.1038/s41598-023-31764-9.
43.
Teshima et al., Antiviral activity of curcumin and its analogs selected by an artificial intelligence-supported activity prediction system in SARS-CoV-2-infected VeroE6 cells, Natural Product Research, doi:10.1080/14786419.2023.2194647.
44.
Zupin et al., Optimization of Anti-SARS-CoV-2 Treatments Based on Curcumin, Used Alone or Employed as a Photosensitizer, Viruses, doi:10.3390/v14102132.
45.
Leka et al., In vitro antiviral activity against SARS-CoV-2 of common herbal medicinal extracts and their bioactive compounds, Phytotherapy Research, doi:10.1002/ptr.7463.
46.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
47.
Marín-Palma et al., Curcumin Inhibits In Vitro SARS-CoV-2 Infection In Vero E6 Cells through Multiple Antiviral Mechanisms, Molecules, doi:10.3390/molecules26226900.
48.
Bahun et al., Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols, Food Chemistry, doi:10.1016/j.foodchem.2021.131594.
49.
Bormann et al., Turmeric Root and Its Bioactive Ingredient Curcumin Effectively Neutralize SARS-CoV-2 In Vitro, Viruses, doi:10.3390/v13101914.
50.
Guijarro-Real et al., Potential In Vitro Inhibition of Selected Plant Extracts against SARS-CoV-2 Chymotripsin-Like Protease (3CLPro) Activity, Foods, doi:10.3390/foods10071503.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
g.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
h.
Non-structural protein 10 (nsp10) serves as an RNA chaperone and stabilizes conformations of nsp12 and nsp14 in the replicase-transcriptase complex, which synthesizes new viral RNAs. Nsp10 disruption may destabilize replicase-transcriptase complex activity.
i.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
j.
The interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor is a primary method of viral entry, inhibiting this interaction can prevent the virus from attaching to and entering host cells, halting infection at an early stage.
k.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
l.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
m.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
n.
A549-AT is a human lung carcinoma cell line stably transfected with ACE2 and TMPRSS2 receptors. Unlike the parental line, this overexpression ensures stable infection and enhanced viral entry, allowing for the evaluation of antiviral efficacy against various SARS-CoV-2 variants.
o.
293T is a human embryonic kidney cell line that can be engineered for high ACE2 expression and SARS-CoV-2 susceptibility. 293T cells are easily transfected and support high protein expression.
p.
HEK293-hACE2 is a human embryonic kidney cell line with high ACE2 expression and SARS-CoV-2 susceptibility. Cells have been transfected with a plasmid to express the human ACE2 (hACE2) protein.
q.
293T/hACE2/TMPRSS2 is a human embryonic kidney cell line engineered for high ACE2 and TMPRSS2 expression, which mimics key aspects of human infection. 293T/hACE2/TMPRSS2 cells are very susceptible to SARS-CoV-2 infection.
r.
Vero E6 is an African green monkey kidney cell line with low/no ACE2 expression and high SARS-CoV-2 susceptibility. The cell line is easy to maintain and supports robust viral replication, however the monkey origin may not accurately represent human responses.
s.
SH-SY5Y is a human neuroblastoma cell line that exhibits neuronal phenotypes. It is commonly used as an in vitro model for studying neurotoxicity, neurodegenerative diseases, and neuronal differentiation.
Singh et al., 29 Jan 2024, preprint, 7 authors.
Contact: khushboo.singh@amway.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach
doi:10.21203/rs.3.rs-3888947/v1
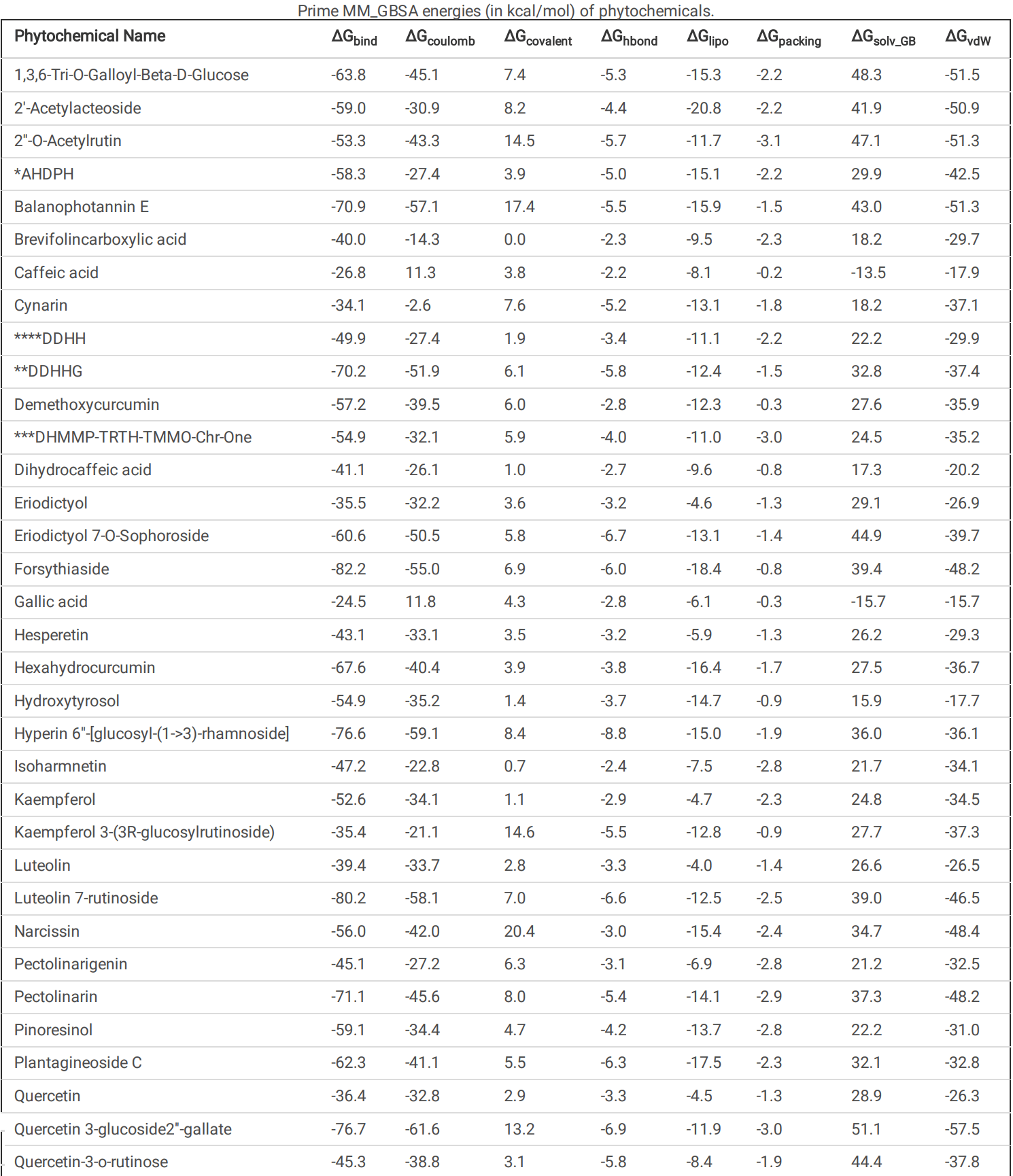
The main protease (M Pro ) of SARS-CoV-2 plays a crucial role in viral replication and is a prime target for therapeutic interventions. Phytochemicals, known for their antiviral properties, have been previously identi ed as potential M Pro inhibitors in several in silico studies. However, the e cacy of these remains in question owing to the inherent exibility of the M Pro binding site, posing challenges in selecting suitable protein structures for virtual screening. In this study, we conducted an extensive analysis of the M Pro binding pocket, utilizing molecular dynamics (MD) simulations to explore its conformational diversity. Based on pocket volume and shape-based clustering, ve representative protein conformations were selected for virtual screening. Virtual screening of a library of ~ 48,000 phytochemicals suggested 39 phytochemicals as potential M Pro inhibitors. Based on subsequent MM-GBSA binding energy calculations and ADMET property predictions, ve compounds were advanced to cell-based viral replication inhibition assays, with three compounds (demethoxycurcumin, shikonin, and withaferin A) exhibiting signi cant (EC50 < 10 uM) inhibition of SARS-CoV-2 replication. Our study provides an understanding of the binding interactions between these phytochemicals and M Pro , contributing signi cantly to the identi cation of promising M Pro inhibitors. Furthermore, beyond its impact on therapeutic development against SARS-CoV-2, this research highlights a crucial role of proper nutrition in the ght against viral infections. Phytochemical Name Docking scores Conformation 1 Conformation 2 Conformation 3 Conformation 4 Conformation 5 1,3,6-Tri-O-Galloyl-Beta-D-Glucose -7.6 -8.8 -11.1 -7.5 -10.1 2'-Acetylacteoside -8.6 -9.5 -12.1 -13.4 -7.6 2''-O-Acetylrutin -10.3 -9.6 -12.2 -10.8 -10.4 *AHDPH -8.1 -9.0 -11.6 -7.5 -9.0 Balanophotannin E -7.5 -11.0 -12.9 -9.9 -8.4 **DDHHG -9.7 -8.3 -10.9 -11.3 -8.6 ***DHMMP-TRTH-TMMO-Chr-One -9.7 -10.5 -10.7 -9.1 -9.9 Eriodictyol 7-O-Sophoroside -12.6 -9.3 -10.0 -11.1 -10.0 Forsythiaside -10.3 -12.6 -14.3 -14.6 -9.2 Hyperin 6''-[glucosyl-(1->3)-rhamnoside] -9.7 -10.9 -15.9 -12.1 -11.9 Kaempferol 3-(3R-glucosylrutinoside) -10.0 -10.6 -12.0 -11.1 -8.5 Luteolin 7-rutinoside -9.8 -9.4 -14.4 -12.0 -9.9 Narcissin -9.7 -10.5 -10.7 -9.1 -9.9 Pectolinarin -8.9 -7.7 -13.9 -8.5 -7.5 Plantagineoside C -9.4 -10.3 -13.3 -10.4 -9.3 Quercetin 3-glucoside2''-gallate -7.8 -9.2 -12.1 -10.6 -7.5 Quercetin-3-o-rutinose -12.2 -11.0 -11.1 -11.5 -11.4 Salvianolic Acid L (SAL) -9.1 -8.2 -13.3 -11.3 -7.6 Shikonin -8.1 -8.4 -8.6 -8.9 -9.5 Shimobashiric Acid C (SAC) -8.2 -8.7 -10.5 -9.6 -10. 2 *AHDPH = (3R,5R)-3-Acetoxy-5-Hydroxy-1,7-Bis(3,4-Dihydroxyphenyl)Heptane. **DDHHG = (3R,5R)-3,5-Dihydroxy-1-(3,4-Dihydroxyphenyl)-7-(4-Hydroxyphenyl)-Heptane 3-O-Beta-D-Glucopyranoside.
For a comprehensive understanding of the model speci cations, validation, and performance, please refer to the AP11.0 user manual and relevant publications 74, 75 .
Cytoxicity Assay Vero cells were seeded using a multiDrop combi liquid dispenser (Thermo) into 384-well plates at a density of 500 cells/well suspended in 50 µL of media. Cells were allowed to recover and fully attach overnight (approximately 16 hours), at which point library compounds were dispensed into wells using an Echo 550 acoustic dispenser (Labcyte). A total of six nal concentrations where tested (50 µM, 25 µM, 12.5 µM, 6.25 µM, 3.125 µM, and 1.5625 µM) and wells were back lled with DMSO such that all wells contained a xed ratio of DMSO. Compounds were incubated with cells for 1 hour prior to addition of virus and then for an additional 24 hours, then xed with 10% formalin, permeabilized 0.1% Triton X-100, washed, and stained for SARS-CoV-2 N protein using a speci c antibody (Sino Biological # MM05) and uorescently labelled secondary antibody. Cells were counter stained with Hoechst 33342 to detect cell nuclei, washed, and imaged with a Cytation 1 (Biotek) automated. Each image was then analyzed with a custom work ow in Cell Pro ler (Broad Inst., Boston, MA) which involved counting of cell nuclei and infected cells. At least 4 replicates were used to construct dose response curves.
Statistics and data normalization The rate index is calculated from cell counts using the following formula: Where X c..
References
Abdusalam, Murugaiyah, Identi cation of Potential Inhibitors of 3CL Protease of SARS-CoV-2 From ZINC Database by Molecular Docking-Based Virtual Screening, Front. Mol. Biosci
Abraham, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX
Agrawal, Blunden, Phytochemicals Against SARS-COV-2 Infection, Nat. Prod. Commun
Alici, Tahtaci, Demir, Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes, Comput. Biol. Chem
Anand, Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain, EMBO J
Anand, Ziebuhr, Wadhwani, Mesters, Hilgenfeld, Coronavirus main proteinase (3CLpro) structure: basis for the design of anti-SARS drugs, Science
Banks, Integrated modeling program applied chemical theory (IMPACT), J. Comp. Chem
Berendsen, Postma, Van Gunsteren, Dinola, Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys
Bharadwaj, Macrolactin A as a Novel Inhibitory Agent for SARS-CoV-2 M pro : Bioinformatics Approach, Appl. Biochem. Biotechnol
Biancatelli, Berrill, Catravas, Marik, Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19), Front. Immunol
Bzówka, Mitusińska, Raczyńska, Samol, Tuszyński et al., Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design, Int. J. Mol. Sci
Cappelli, Manganelli, Lombardo, Gissi, Benfenati, Validation of quantitative structure-activity relationship models to predict water-solubility of organic compounds, Sci. Total Environ
Chakraborty, The Natural Products Withaferin A and Withanone from the Medicinal Herb Withania somnifera Are Covalent Inhibitors of the SARS-CoV-2 Main Protease, J. Nat. Prod
Cherrak, Merzouk, Mokhtari-Soulimane, Potential bioactive glycosylated avonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies, PLoS One
Da Fonseca, Screening of Potential Inhibitors Targeting the Main Protease Structure of SARS-CoV-2 via Molecular Docking, and Approach with Molecular Dynamics, RMSD, RMSF, H-Bond, SASA and MMGBSA, Mol. Biotechnol, doi:Preprintat10.1007/s12033-023-00831-x
Dai, Zhang, Jiang, Su, Li, Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease, Science
Darden, York, Pedersen, Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems, J. Chem. Phys
Dearden, In silico prediction of aqueous solubility, Expet Opin. Drug Discov
Dhawan, Anti-viral activity of Indian plants, Proc. Natl. Acad. Sci. India Sect. B Biol. Sci
Douangamath, Fearon, Gehrtz, Krojer, Lukacik, Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease, Nat. Commun
Durdagi, Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing, Structure
Durrant, POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics, J. Chem. Theory Comput
Estrada, Topological analysis of SARS CoV-2 main protease, Chaos
Fei, Contribution of traditional Chinese medicine combined with conventional western medicine treatment for the novel coronavirus disease (COVID-19), current evidence with systematic review and meta-analysis, Phytother. Res
Flynn, Comprehensive tness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms, Elife
Galan, Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-COV-2 infection, Pathog. Glob. Health
Ghosh, Structure-activity relationship (SAR) and molecular dynamics study of withaferin-A fragment derivatives as a potential therapeutic lead against the main protease (Mpro) of SARS-CoV-2, J. Mol. Model
Gorbalenya, The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-ncov and naming it SARS-COV-2, Nature Microbiol
Gossen, A Blueprint for High A nity SARS-CoV-2 Mpro Inhibitors from Activity-Based Compound Library Screening Guided by Analysis of Protein Dynamics, ACS Pharmacol. Transl. Sci
Gupta, Identi cation of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies, J. Biomol. Struct. Dyn
Gupta, Structure-Based Virtual Screening and Biochemical Validation to Discover Potential Inhibitor of the SARS-CoV-2 Main Protease, ACS Omega
Henrich, Beutler, Matching the power of high throughput screening to the chemical diversity of natural products, Nat. Prod. Rep
Hess, Bekker, Berendsen, Fraaije, Lincs, A linear constraint solver for molecular simulations, J. Comp. Chem
Huang, Mackerell, CHARMM36 all-atom additive protein force eld: validation based on comparison to NMR data, J. Comput. Chem
Humphrey, Dalke, Schulten, VMD -Visual Molecular Dynamics, J. Mol. Graphics
Issa, The Main Protease of SARS-CoV-2 as a Target for Phytochemicals against Coronavirus, Plants
Jamhour, Phytochemicals As a Potential Inhibitor of COVID-19: An In-Silico Perspective, Russ. J. Phys. Chem
Jin, Structure of M pro from SARS-CoV-2 and discovery of its inhibitors, Nature
Jorgensen, Chandrasekhar, Madura, Impey, Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys
Kaur, How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions, Physiol. Mol. Biol. Plants
Khaerunnisa, Kurniawan, Awaluddin, Suhartati, Soetjipto, Potential Inhibitor of COVID-19 Main Protease (M pro ) From Several Medicinal Plant Compounds by Molecular Docking Study, doi:10.20944/preprints202003.0226.v1
Khanna, Herbal Immune-boosters: Substantial warriors of pandemiccovid-19 battle, Phytomedicine
Kneller, Kovalevsky, Coates, Structural plasticity of the SARS-COV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography, Nature Commun
Lachance, Charting, navigating, and populating natural product chemical space for drug discovery, J. Med. Chem
Lawson, Maccoss, Heer, Importance of rigidity in designing small molecule drugs to tackle protein-protein interactions (ppis) through stabilization of desired conformers, J. Med. Chem
Li, Abel, Zhu, Cao, Zhao et al., The VSGB 2.0 model: a next-generation energy model for high-resolution protein structure modeling, Proteins
Ling, Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-COV-2), J. Integr. Med
Ma, Disul ram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspeci c Promiscuous SARS-CoV-2 Main Protease Inhibitors, ACS Pharmacol. Transl. Sci
Mani, Natural product-derived phytochemicals as potential agents against coronaviruses: a review, Virus Res
Mulu, The impact of curcumin-derived polyphenols on the structure and exibility COVID-19 main protease binding pocket: a molecular dynamics simulation study, PeerJ
Pettersen, UCSF Chimera -A visualization system for exploratory research and analysis, J. Comp. Chem
Remali, Aizat, A review on plant bioactive compounds and their modes of action against coronavirus infection, Front. Pharmacol
Ren, The newly emerged SARS-like coronavirus HCoV-EMC also has an "Achilles' heel": current effective inhibitor targeting a 3C-like protease, Protein Cell
Romano, Tatonetti, Informatics and computational methods in natural product drug discovery: A review and Perspectives, Front. Genet
Singh, Briggs, Impact of lymphoma-linked Asn11Tyr point mutation on the interaction between Bcl-2 and a BH3 mimetic: Insights from molecular dynamics simulation, Chem. Biol. Drug Design
Sztain, Amaro, Mccammon, Elucidation of cryptic and allosteric pockets within the SARS-CoV-2 protease, J. Chem. Inf. Model
Teli, Fragment-based design of SARS-CoV-2 Mpro inhibitors, Struct Chem
Vallejos, Ivermectin to prevent hospitalizations in patients with covid-19 (IVERCOR-covid19) a randomized, double-blind, placebocontrolled trial, BMC Infect. Dis
Wagner, POVME 3.0: Software for Mapping Binding Pocket Flexibility, J. Chem. Theory Comput
Wang, Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design, Sci. Rep
Wu, A new coronavirus associated with human respiratory disease in China, Nature
Wu, Author Correction: A New Coronavirus Associated with Human Respiratory Disease in China, Nature
Xue, Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design, J. Virol
Yang, Screening of potential inhibitors targeting the main protease structure of SARS-CoV-2 via molecular docking, Front. Pharmacol
Yang, The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor, Proc. Natl. Acad. Sci
Zeng, CMAUP: A database of collective molecular activities of useful plants, Nucleic Acids Res
Zhang, Hilgenfeld, Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Glnvinylsulfone, doi:10.2210/pdb6Z2E/pdb
Zhang, Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus, J. Virol
Zhou, A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature
DOI record:
{
"DOI": "10.21203/rs.3.rs-3888947/v1",
"URL": "http://dx.doi.org/10.21203/rs.3.rs-3888947/v1",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The main protease (M<jats:sup>Pro</jats:sup>) of SARS-CoV-2 plays a crucial role in viral replication and is a prime target for therapeutic interventions. Phytochemicals, known for their antiviral properties, have been previously identified as potential M<jats:sup>Pro</jats:sup> inhibitors in several in silico studies. However, the efficacy of these remains in question owing to the inherent flexibility of the M<jats:sup>Pro</jats:sup> binding site, posing challenges in selecting suitable protein structures for virtual screening. In this study, we conducted an extensive analysis of the M<jats:sup>Pro</jats:sup> binding pocket, utilizing molecular dynamics (MD) simulations to explore its conformational diversity. Based on pocket volume and shape-based clustering, five representative protein conformations were selected for virtual screening. Virtual screening of a library of ~ 48,000 phytochemicals suggested 39 phytochemicals as potential M<jats:sup>Pro</jats:sup> inhibitors. Based on subsequent MM-GBSA binding energy calculations and ADMET property predictions, five compounds were advanced to cell-based viral replication inhibition assays, with three compounds (demethoxycurcumin, shikonin, and withaferin A) exhibiting significant (EC50 < 10 uM) inhibition of SARS-CoV-2 replication. Our study provides an understanding of the binding interactions between these phytochemicals and M<jats:sup>Pro</jats:sup>, contributing significantly to the identification of promising M<jats:sup>Pro</jats:sup> inhibitors. Furthermore, beyond its impact on therapeutic development against SARS-CoV-2, this research highlights a crucial role of proper nutrition in the fight against viral infections.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
1,
22
]
]
},
"author": [
{
"affiliation": [
{
"name": "Amway (United States)"
}
],
"family": "Singh",
"given": "Khushboo",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Boston University"
}
],
"family": "Patten",
"given": "J. J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "The University of Texas Medical Branch at Galveston"
}
],
"family": "Dimet",
"given": "Andrea",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Boston University"
}
],
"family": "Davey",
"given": "Robert A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "The University of Texas Medical Branch at Galveston"
}
],
"family": "Watowich",
"given": "Stanley J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Amway (United States)"
}
],
"family": "Chandra",
"given": "Amit",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Amway (United States)"
}
],
"family": "Leverett",
"given": "Jesse",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
1,
29
]
],
"date-time": "2024-01-29T10:28:18Z",
"timestamp": 1706524098000
},
"deposited": {
"date-parts": [
[
2024,
1,
29
]
],
"date-time": "2024-01-29T10:28:52Z",
"timestamp": 1706524132000
},
"group-title": "In Review",
"indexed": {
"date-parts": [
[
2024,
1,
30
]
],
"date-time": "2024-01-30T00:22:24Z",
"timestamp": 1706574144551
},
"institution": [
{
"name": "Research Square"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
1,
29
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
1,
29
]
],
"date-time": "2024-01-29T00:00:00Z",
"timestamp": 1706486400000
}
}
],
"link": [
{
"URL": "https://www.researchsquare.com/article/rs-3888947/v1",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.researchsquare.com/article/rs-3888947/v1.html",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "8761",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
1,
29
]
]
},
"prefix": "10.21203",
"published": {
"date-parts": [
[
2024,
1,
29
]
]
},
"publisher": "Research Square Platform LLC",
"reference": [
{
"DOI": "10.1038/s41586-020-2202-3",
"article-title": "Author Correction: A New Coronavirus Associated with Human Respiratory Disease in China",
"author": "Wu F",
"doi-asserted-by": "crossref",
"first-page": "E7",
"journal-title": "Nature.",
"key": "ref1",
"unstructured": "Wu, F., et al. Author Correction: A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 580, E7 (2020).",
"volume": "580",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"article-title": "The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-ncov and naming it SARS-COV-2",
"author": "Gorbalenya AE",
"doi-asserted-by": "crossref",
"first-page": "536",
"journal-title": "Nature Microbiol",
"key": "ref2",
"unstructured": "Gorbalenya, A.E. et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-ncov and naming it SARS-COV-2. Nature Microbiol. 5, 536–544 (2020).",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1080/20477724.2021.1890887",
"article-title": "Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-COV-2 infection",
"author": "Galan LE",
"doi-asserted-by": "crossref",
"first-page": "235",
"journal-title": "Pathog. Glob. Health.",
"key": "ref3",
"unstructured": "Galan, L.E. et al. Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-COV-2 infection. Pathog. Glob. Health. 115, 235–242 (2021).",
"volume": "115",
"year": "2021"
},
{
"DOI": "10.1186/s12879-021-06348-5",
"article-title": "Ivermectin to prevent hospitalizations in patients with covid-19 (IVERCOR-covid19) a randomized, double-blind, placebo-controlled trial",
"author": "Vallejos J",
"doi-asserted-by": "crossref",
"first-page": "635",
"journal-title": "BMC Infect. Dis",
"key": "ref4",
"unstructured": "Vallejos, J. et al. Ivermectin to prevent hospitalizations in patients with covid-19 (IVERCOR-covid19) a randomized, double-blind, placebo-controlled trial. BMC Infect. Dis. 21, 635 (2021)",
"volume": "21",
"year": "2021"
},
{
"author": "Dhawan BN",
"key": "ref5",
"unstructured": "Dhawan, B.N. Anti-viral activity of Indian plants. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 82, 209–224 (2012).",
"year": "2012"
},
{
"article-title": "Phytochemical, antioxidant and microbial inhibitory effects of spondias mombin leaf and stem bark extracts",
"first-page": "14",
"journal-title": "IOSR J. Pharm. Biol. Sci.",
"key": "ref6",
"unstructured": "H.C.C, M. et al. Phytochemical, antioxidant and microbial inhibitory effects of spondias mombin leaf and stem bark extracts. IOSR J. Pharm. Biol. Sci. 9, pp. 14–17 (2014).",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1016/j.phymed.2020.153361",
"article-title": "Herbal Immune-boosters: Substantial warriors of pandemiccovid-19 battle",
"author": "Khanna K",
"doi-asserted-by": "crossref",
"first-page": "153361",
"journal-title": "Phytomedicine",
"key": "ref7",
"unstructured": "Khanna, K. et al. Herbal Immune-boosters: Substantial warriors of pandemiccovid-19 battle. Phytomedicine. 85, 153361 (2021).",
"volume": "85",
"year": "2021"
},
{
"DOI": "10.1016/j.joim.2020.02.004",
"article-title": "Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-COV-2)",
"author": "Ling C",
"doi-asserted-by": "crossref",
"first-page": "8788",
"journal-title": "J. Integr. Med.",
"key": "ref8",
"unstructured": "Ling, C. Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-COV-2). J. Integr. Med. 18, 8788 (2020).",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.3389/fphar.2020.589044",
"article-title": "A review on plant bioactive compounds and their modes of action against coronavirus infection",
"author": "Remali J",
"doi-asserted-by": "crossref",
"first-page": "589044",
"journal-title": "Front. Pharmacol.",
"key": "ref9",
"unstructured": "Remali, J. & Aizat, W.M. A review on plant bioactive compounds and their modes of action against coronavirus infection. Front. Pharmacol. 11, 589044 (2021).",
"volume": "11",
"year": "2021"
},
{
"article-title": "Contribution of traditional Chinese medicine combined with conventional western medicine treatment for the novel coronavirus disease (COVID-19), current evidence with systematic review and meta‐analysis",
"author": "Fei J",
"journal-title": "Phytother. Res.",
"key": "ref10",
"unstructured": "Fei, J. et al. Contribution of traditional Chinese medicine combined with conventional western medicine treatment for the novel coronavirus disease (COVID-19), current evidence with systematic review and meta‐analysis. Phytother. Res. 35, (2021).",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1021/acs.jmedchem.7b01120",
"article-title": "Importance of rigidity in designing small molecule drugs to tackle protein–protein interactions (ppis) through stabilization of desired conformers",
"author": "Lawson AD",
"doi-asserted-by": "crossref",
"first-page": "4283",
"journal-title": "J. Med. Chem.",
"key": "ref11",
"unstructured": "Lawson, A.D., MacCoss, M. & Heer, J.P. Importance of rigidity in designing small molecule drugs to tackle protein–protein interactions (ppis) through stabilization of desired conformers. J. Med. Chem. 61, 4283–4289 (2017).",
"volume": "61",
"year": "2017"
},
{
"DOI": "10.1039/c3np70052f",
"article-title": "Matching the power of high throughput screening to the chemical diversity of natural products",
"author": "Henrich CJ",
"doi-asserted-by": "crossref",
"first-page": "1284",
"journal-title": "Nat. Prod. Rep.",
"key": "ref12",
"unstructured": "Henrich, C.J. & Beutler, J.A. Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 30, 1284 (2013).",
"volume": "30",
"year": "2013"
},
{
"DOI": "10.3389/fgene.2019.00368",
"article-title": "Informatics and computational methods in natural product drug discovery: A review and Perspectives",
"author": "Romano JD",
"doi-asserted-by": "crossref",
"first-page": "368",
"journal-title": "Front. Genet.",
"key": "ref13",
"unstructured": "Romano, J.D. & Tatonetti, N.P. Informatics and computational methods in natural product drug discovery: A review and Perspectives. Front. Genet. 10, 368 (2019).",
"volume": "10",
"year": "2019"
},
{
"article-title": "A database of collective molecular activities of useful plants",
"author": "Zeng X",
"first-page": "D1118-D1127",
"journal-title": "Nucleic Acids Res",
"key": "ref14",
"unstructured": "Zeng, X. et al. CMAUP: A database of collective molecular activities of useful plants. Nucleic Acids Res. 47, D1118-D1127 (2018).",
"volume": "CMAUP",
"year": "2018"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A pneumonia outbreak associated with a new coronavirus of probable bat origin",
"author": "Zhou P",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nature",
"key": "ref15",
"unstructured": "Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273 (2020).",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"article-title": "A new coronavirus associated with human respiratory disease in China",
"author": "Wu F",
"doi-asserted-by": "crossref",
"first-page": "265",
"journal-title": "Nature.",
"key": "ref16",
"unstructured": "Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature. 579, 265–269 (2020).",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1093/emboj/cdf327",
"article-title": "Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain",
"author": "Anand K",
"doi-asserted-by": "crossref",
"first-page": "3213",
"journal-title": "EMBO J.",
"key": "ref17",
"unstructured": "Anand, K. et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 21, 3213–3224 (2002).",
"volume": "21",
"year": "2002"
},
{
"author": "Yang H",
"key": "ref18",
"unstructured": "Yang, H. et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 100, 13190–13195 (2003).",
"year": "2003"
},
{
"DOI": "10.1128/JVI.02114-07",
"article-title": "Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design",
"author": "Xue X",
"doi-asserted-by": "crossref",
"first-page": "2515",
"journal-title": "J. Virol.",
"key": "ref19",
"unstructured": "Xue, X. et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82, 2515–2527 (2008).",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1007/s13238-013-2841-3",
"article-title": "The newly emerged SARS-like coronavirus HCoV-EMC also has an \"Achilles' heel\": current effective inhibitor targeting a 3C-like protease",
"author": "Ren Z",
"doi-asserted-by": "crossref",
"first-page": "248",
"journal-title": "Protein Cell.",
"key": "ref20",
"unstructured": "Ren, Z. et al. The newly emerged SARS-like coronavirus HCoV-EMC also has an \"Achilles' heel\": current effective inhibitor targeting a 3C-like protease. Protein Cell. 4, 248–250 (2013).",
"volume": "4",
"year": "2013"
},
{
"DOI": "10.1038/srep22677",
"article-title": "Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design",
"author": "Wang F",
"doi-asserted-by": "crossref",
"first-page": "22677",
"journal-title": "Sci. Rep.",
"key": "ref21",
"unstructured": "Wang, F. et al. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 6, 22677 (2016).",
"volume": "6",
"year": "2016"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors.",
"author": "Jin Z",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "ref22",
"unstructured": "Jin, Z. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 582, 289–293 (2020).",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-16954-7",
"article-title": "Structural plasticity of the SARS-COV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography",
"author": "Kneller DW",
"doi-asserted-by": "crossref",
"first-page": "3202",
"journal-title": "Nature Commun",
"key": "ref23",
"unstructured": "Kneller, D.W., Kovalevsky, A. & Coates, L. Structural plasticity of the SARS-COV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nature Commun. 11, 3202 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3390/ijms21093099",
"article-title": "Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design",
"author": "Bzówka M",
"doi-asserted-by": "crossref",
"first-page": "3099",
"journal-title": "Int. J. Mol. Sci.",
"key": "ref24",
"unstructured": "Bzówka, M., Mitusińska, K., Raczyńska, A., Samol, A., Tuszyński, J.A., & Góra, A. Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design. Int. J. Mol. Sci. 21, 3099 (2020).",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1021/acsptsci.0c00215",
"article-title": "A Blueprint for High Affinity SARS-CoV-2 Mpro Inhibitors from Activity-Based Compound Library Screening Guided by Analysis of Protein Dynamics",
"author": "Gossen J",
"doi-asserted-by": "crossref",
"first-page": "1079",
"journal-title": "ACS Pharmacol. Transl. Sci.",
"key": "ref25",
"unstructured": "Gossen, J. et al. A Blueprint for High Affinity SARS-CoV-2 Mpro Inhibitors from Activity-Based Compound Library Screening Guided by Analysis of Protein Dynamics. ACS Pharmacol. Transl. Sci. 4, 1079–1095 (2021).",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.7554/eLife.77433",
"article-title": "Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms",
"author": "Flynn JM",
"doi-asserted-by": "crossref",
"first-page": "e77433",
"journal-title": "Elife.",
"key": "ref26",
"unstructured": "Flynn, J.M. et al. Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms. Elife. 11, e77433 (2022).",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.1063/5.0013029",
"article-title": "Topological analysis of SARS CoV-2 main protease",
"author": "Estrada E",
"doi-asserted-by": "crossref",
"first-page": "061102",
"journal-title": "Chaos",
"key": "ref27",
"unstructured": "Estrada, E. Topological analysis of SARS CoV-2 main protease. Chaos. 30, 061102 (2020).",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1007/s12010-021-03608-7",
"article-title": "Macrolactin A as a Novel Inhibitory Agent for SARS-CoV-2 Mpro: Bioinformatics Approach",
"author": "Bharadwaj KK",
"doi-asserted-by": "crossref",
"first-page": "3371",
"journal-title": "Appl. Biochem. Biotechnol.",
"key": "ref28",
"unstructured": "Bharadwaj, K.K. et al. Macrolactin A as a Novel Inhibitory Agent for SARS-CoV-2 Mpro: Bioinformatics Approach. Appl. Biochem. Biotechnol. 193, 3371–3394 (2021).",
"volume": "193",
"year": "2021"
},
{
"DOI": "10.3389/fmolb.2020.603037",
"article-title": "Identification of Potential Inhibitors of 3CL Protease of SARS-CoV-2 From ZINC Database by Molecular Docking-Based Virtual Screening",
"author": "Abdusalam AAA",
"doi-asserted-by": "crossref",
"first-page": "603037",
"journal-title": "Front. Mol. Biosci.",
"key": "ref29",
"unstructured": "Abdusalam, A.A.A., & Murugaiyah, V. Identification of Potential Inhibitors of 3CL Protease of SARS-CoV-2 From ZINC Database by Molecular Docking-Based Virtual Screening. Front. Mol. Biosci. 7, 603037 (2020).",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1021/acsomega.0c04808",
"article-title": "Structure-Based Virtual Screening and Biochemical Validation to Discover a Potential Inhibitor of the SARS-CoV-2 Main Protease",
"author": "Gupta A",
"doi-asserted-by": "crossref",
"first-page": "33151",
"journal-title": "ACS Omega",
"key": "ref30",
"unstructured": "Gupta, A. et al. Structure-Based Virtual Screening and Biochemical Validation to Discover a Potential Inhibitor of the SARS-CoV-2 Main Protease. ACS Omega. 5, 33151–33161 (2020).",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1021/ct500381c",
"article-title": "POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics",
"author": "Durrant JD",
"doi-asserted-by": "crossref",
"first-page": "5047",
"journal-title": "J. Chem. Theory Comput",
"key": "ref31",
"unstructured": "Durrant, J.D. et al. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J. Chem. Theory Comput. 10, 5047–5056 (2014).",
"volume": "10",
"year": "2014"
},
{
"DOI": "10.1021/acs.jctc.7b00500",
"article-title": "POVME 3.0: Software for Mapping Binding Pocket Flexibility",
"author": "Wagner JR",
"doi-asserted-by": "crossref",
"first-page": "4584",
"journal-title": "J. Chem. Theory Comput.",
"key": "ref32",
"unstructured": "Wagner, J.R. et al. POVME 3.0: Software for Mapping Binding Pocket Flexibility. J. Chem. Theory Comput. 13, 4584–4592 (2017).",
"volume": "13",
"year": "2017"
},
{
"DOI": "10.1016/j.str.2021.07.007",
"article-title": "Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing",
"author": "Durdagi S",
"doi-asserted-by": "crossref",
"first-page": "1382",
"journal-title": "Structure.",
"key": "ref33",
"unstructured": "Durdagi, S. et al. Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing. Structure. 29, 1382–1396 (2021).",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.1021/acs.jcim.1c00140",
"article-title": "Elucidation of cryptic and allosteric pockets within the SARS-CoV-2 protease",
"author": "Sztain T",
"doi-asserted-by": "crossref",
"first-page": "3495",
"journal-title": "J. Chem. Inf. Model",
"key": "ref34",
"unstructured": "Sztain, T., Amaro, R., & McCammon, J.A. Elucidation of cryptic and allosteric pockets within the SARS-CoV-2 protease. J. Chem. Inf. Model. 61, 3495–3501 (2021).",
"volume": "61",
"year": "2021"
},
{
"DOI": "10.3389/fphar.2022.962863",
"article-title": "Screening of potential inhibitors targeting the main protease structure of SARS-CoV-2 via molecular docking",
"author": "Yang X",
"doi-asserted-by": "crossref",
"first-page": "962863",
"journal-title": "Front. Pharmacol.",
"key": "ref35",
"unstructured": "Yang, X. et al. Screening of potential inhibitors targeting the main protease structure of SARS-CoV-2 via molecular docking. Front. Pharmacol. 13, 962863 (2022).",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1007/s12033-023-00831-x",
"author": "Fonseca AM",
"doi-asserted-by": "publisher",
"key": "ref36",
"unstructured": "da Fonseca, A.M. et al. Screening of Potential Inhibitors Targeting the Main Protease Structure of SARS-CoV-2 via Molecular Docking, and Approach with Molecular Dynamics, RMSD, RMSF, H-Bond, SASA and MMGBSA. Mol. Biotechnol. Preprint at 10.1007/s12033-023-00831-x (2023).",
"year": "2023"
},
{
"DOI": "10.1007/s11224-022-02031-w",
"article-title": "Fragment-based design of SARS-CoV-2 Mpro inhibitors",
"author": "Teli DM",
"doi-asserted-by": "crossref",
"first-page": "2155",
"journal-title": "Struct Chem",
"key": "ref37",
"unstructured": "Teli, D.M. et al. Fragment-based design of SARS-CoV-2 Mpro inhibitors. Struct Chem. 33, 2155–2168 (2022).",
"volume": "33",
"year": "2022"
},
{
"DOI": "10.1134/S0036024422070251",
"article-title": "Phytochemicals As a Potential Inhibitor of COVID-19: An In-Silico Perspective",
"author": "Jamhour RMAQ",
"doi-asserted-by": "crossref",
"first-page": "1589",
"journal-title": "Russ. J. Phys. Chem.",
"key": "ref38",
"unstructured": "Jamhour, R.M.A.Q. et al. Phytochemicals As a Potential Inhibitor of COVID-19: An In-Silico Perspective. Russ. J. Phys. Chem. 96, 1589–97 (2022).",
"volume": "96",
"year": "2022"
},
{
"article-title": "Phytochemicals Against SARS-COV-2 Infection",
"author": "Agrawal PK",
"journal-title": "Nat. Prod. Commun.",
"key": "ref39",
"unstructured": "Agrawal, P.K., & Blunden, G. Phytochemicals Against SARS-COV-2 Infection. Nat. Prod. Commun. 18, (2023).",
"volume": "18",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2020.01451",
"article-title": "Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19)",
"author": "Biancatelli RMLC",
"doi-asserted-by": "crossref",
"first-page": "1451",
"journal-title": "Front. Immunol.",
"key": "ref40",
"unstructured": "Biancatelli, R.M.L.C., Berrill, M., Catravas, J., & Marik, P.E. Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front. Immunol. 11, 1451 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.virusres.2020.197989",
"article-title": "Natural product-derived phytochemicals as potential agents against coronaviruses: a review",
"author": "Mani JS",
"doi-asserted-by": "crossref",
"first-page": "197989",
"journal-title": "Virus Res.",
"key": "ref41",
"unstructured": "Mani J.S et al. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 284, 197989. (2020).",
"volume": "284",
"year": "2020"
},
{
"DOI": "10.3390/plants11141862",
"article-title": "The Main Protease of SARS-CoV-2 as a Target for Phytochemicals against Coronavirus",
"author": "Issa SS",
"doi-asserted-by": "crossref",
"first-page": "1862",
"journal-title": "Plants",
"key": "ref42",
"unstructured": "Issa S.S. et al. The Main Protease of SARS-CoV-2 as a Target for Phytochemicals against Coronavirus. Plants. 11, 1862 (2022).",
"volume": "11",
"year": "2022"
},
{
"key": "ref43",
"unstructured": "AP11.0, SimulationsPlus LLC, Lancaster, CA."
},
{
"DOI": "10.1016/j.compbiolchem.2022.107657",
"article-title": "Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes",
"author": "Alici H",
"doi-asserted-by": "crossref",
"journal-title": "Comput. Biol. Chem.",
"key": "ref44",
"unstructured": "Alici, H., Tahtaci, H., & Demir, K. Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes. Comput. Biol. Chem. (2022).",
"year": "2022"
},
{
"DOI": "10.7717/peerj.11590",
"article-title": "The impact of curcumin-derived polyphenols on the structure and flexibility COVID-19 main protease binding pocket: a molecular dynamics simulation study",
"author": "Mulu A",
"doi-asserted-by": "crossref",
"first-page": "e11590",
"journal-title": "PeerJ",
"key": "ref45",
"unstructured": "Mulu, A. et al. The impact of curcumin-derived polyphenols on the structure and flexibility COVID-19 main protease binding pocket: a molecular dynamics simulation study. PeerJ. 9, e11590 (2021).",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.20944/preprints202003.0226.v1",
"author": "Khaerunnisa S",
"doi-asserted-by": "publisher",
"key": "ref46",
"unstructured": "Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., & Soetjipto, S. Potential Inhibitor of COVID-19 Main Protease (Mpro) From Several Medicinal Plant Compounds by Molecular Docking Study. Preprint at https://doi.org/10.20944/preprints202003.0226.v1 (2020).",
"year": "2020"
},
{
"DOI": "10.1007/s00894-021-04703-6",
"article-title": "Structure-activity relationship (SAR) and molecular dynamics study of withaferin-A fragment derivatives as a potential therapeutic lead against the main protease (Mpro) of SARS-CoV-2",
"author": "Ghosh A",
"doi-asserted-by": "crossref",
"first-page": "97",
"journal-title": "J. Mol. Model.",
"key": "ref47",
"unstructured": "Ghosh, A. et al. Structure-activity relationship (SAR) and molecular dynamics study of withaferin-A fragment derivatives as a potential therapeutic lead against the main protease (Mpro) of SARS-CoV-2. J. Mol. Model. 27, 97 (2021).",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1021/acs.jnatprod.2c00521",
"article-title": "The Natural Products Withaferin A and Withanone from the Medicinal Herb Withania somnifera Are Covalent Inhibitors of the SARS-CoV-2 Main Protease",
"author": "Chakraborty S",
"doi-asserted-by": "crossref",
"first-page": "2340",
"journal-title": "J. Nat. Prod.",
"key": "ref48",
"unstructured": "Chakraborty, S. et al. The Natural Products Withaferin A and Withanone from the Medicinal Herb Withania somnifera Are Covalent Inhibitors of the SARS-CoV-2 Main Protease. J. Nat. Prod. 85, 2340–2350 (2022).",
"volume": "85",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01253-21",
"article-title": "Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus",
"author": "Zhang Y",
"doi-asserted-by": "crossref",
"first-page": "e0125321",
"journal-title": "J. Virol.",
"key": "ref49",
"unstructured": "Zhang, Y. et al. Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus. J. Virol. 96, e0125321 (2022).",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1021/acsptsci.0c00130",
"article-title": "PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors",
"author": "Ma C",
"doi-asserted-by": "crossref",
"first-page": "1265",
"journal-title": "ACS Pharmacol. Transl. Sci.",
"key": "ref50",
"unstructured": "Ma, C. et al. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 3, 1265–1277 (2020).",
"volume": "3",
"year": "2020"
},
{
"DOI": "10.1080/07391102.2020.1776157",
"article-title": "Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies",
"author": "Gupta S",
"doi-asserted-by": "crossref",
"first-page": "4334",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "ref51",
"unstructured": "Gupta, S. et al. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 39, 4334–4345 (2021).",
"volume": "39",
"year": "2021"
},
{
"DOI": "10.1021/jm300288g",
"article-title": "Charting, navigating, and populating natural product chemical space for drug discovery",
"author": "Lachance H",
"doi-asserted-by": "crossref",
"first-page": "5989",
"journal-title": "J. Med. Chem.",
"key": "ref52",
"unstructured": "Lachance, H. et al. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 55, 5989–6001 (2012).",
"volume": "55",
"year": "2012"
},
{
"DOI": "10.1007/s12298-022-01146-y",
"article-title": "How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions",
"author": "Kaur S",
"doi-asserted-by": "crossref",
"first-page": "485",
"journal-title": "Physiol. Mol. Biol. Plants",
"key": "ref53",
"unstructured": "Kaur, S. et al. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants. 28, 485–504 (2022).",
"volume": "28",
"year": "2022"
},
{
"key": "ref54",
"unstructured": "Protein Data Bank. Retrieved August 29, 2023, from https://www.rcsb.org"
},
{
"DOI": "10.2210/pdb6Z2E/pdb",
"article-title": "Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)",
"author": "Zhang L",
"doi-asserted-by": "publisher",
"key": "ref55",
"unstructured": "Zhang, L., & Hilgenfeld, R. Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone. https://doi.org/10.2210/pdb6Z2E/pdb (2020).",
"year": "2020"
},
{
"DOI": "10.1126/science.abb4489",
"article-title": "Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease",
"author": "Dai W",
"doi-asserted-by": "crossref",
"first-page": "1331",
"journal-title": "Science.",
"key": "ref56",
"unstructured": "Dai, W., Zhang, B., Jiang, X.M., Su, H., & Li, J. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 368, 1331–1335 (2020).",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1126/science.1085658",
"article-title": "Coronavirus main proteinase (3CLpro) structure: basis for the design of anti-SARS drugs",
"author": "Anand K",
"doi-asserted-by": "crossref",
"first-page": "1763",
"journal-title": "Science",
"key": "ref57",
"unstructured": "Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J.R., & Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: basis for the design of anti-SARS drugs. Science. 300, 1763–1767 (2020).",
"volume": "300",
"year": "2020"
},
{
"DOI": "10.1371/journal.pone.0240653",
"article-title": "Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies",
"author": "Cherrak SA",
"doi-asserted-by": "crossref",
"first-page": "e0240653",
"journal-title": "PLoS One",
"key": "ref58",
"unstructured": "Cherrak, S.A., Merzouk, H., & Mokhtari-Soulimane, N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS One. 15, e0240653 (2020).",
"volume": "15",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-18709-w",
"article-title": "Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease",
"author": "Douangamath A",
"doi-asserted-by": "crossref",
"first-page": "5047",
"journal-title": "Nat. Commun.",
"key": "ref59",
"unstructured": "Douangamath, A., Fearon, D., Gehrtz, P., Krojer, T., & Lukacik, P. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 11, 5047 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.softx.2015.06.001",
"article-title": "High performance molecular simulations through multi-level parallelism from laptops to supercomputers",
"author": "Abraham MJ",
"doi-asserted-by": "crossref",
"first-page": "19",
"journal-title": "SoftwareX",
"key": "ref60",
"unstructured": "Abraham, M.J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1, 19–25 (2015).",
"volume": "GROMACS",
"year": "2015"
},
{
"DOI": "10.1002/jcc.23354",
"article-title": "CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data",
"author": "Huang J",
"doi-asserted-by": "crossref",
"first-page": "2135",
"journal-title": "J. Comput. Chem.",
"key": "ref61",
"unstructured": "Huang, J., & MacKerell, A.D. Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).",
"volume": "34",
"year": "2013"
},
{
"DOI": "10.1063/1.445869",
"article-title": "Comparison of simple potential functions for simulating liquid water",
"author": "Jorgensen WL",
"doi-asserted-by": "crossref",
"first-page": "926",
"journal-title": "J. Chem. Phys.",
"key": "ref62",
"unstructured": "Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W., & Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 (1983).",
"volume": "79",
"year": "1983"
},
{
"DOI": "10.1063/1.464397",
"article-title": "Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems",
"author": "Darden T",
"doi-asserted-by": "crossref",
"first-page": "10089",
"journal-title": "J. Chem. Phys.",
"key": "ref63",
"unstructured": "Darden, T., York, D., & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).",
"volume": "98",
"year": "1993"
},
{
"DOI": "10.1111/cbdd.13653",
"article-title": "Impact of lymphoma-linked Asn11Tyr point mutation on the interaction between Bcl-2 and a BH3 mimetic: Insights from molecular dynamics simulation",
"author": "Singh K",
"doi-asserted-by": "crossref",
"first-page": "435",
"journal-title": "Chem. Biol. Drug Design.",
"key": "ref64",
"unstructured": "Singh,K. & Briggs, J.M. Impact of lymphoma-linked Asn11Tyr point mutation on the interaction between Bcl-2 and a BH3 mimetic: Insights from molecular dynamics simulation. Chem. Biol. Drug Design. 95, 435–450 (2020).",
"volume": "95",
"year": "2020"
},
{
"DOI": "10.1063/1.448118",
"article-title": "Molecular dynamics with coupling to an external bath",
"author": "Berendsen H",
"doi-asserted-by": "crossref",
"first-page": "3684",
"journal-title": "J. Chem. Phys.",
"key": "ref65",
"unstructured": "Berendsen, H., Postma, J., van Gunsteren, W., DiNola, A., & Haak, J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).",
"volume": "81",
"year": "1984"
},
{
"DOI": "10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H",
"article-title": "E. M. LINCS: A linear constraint solver for molecular simulations",
"author": "Hess B",
"doi-asserted-by": "crossref",
"journal-title": "J. Comp. Chem.",
"key": "ref66",
"unstructured": "Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 18, (1997).",
"volume": "18",
"year": "1997"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"article-title": "VMD - Visual Molecular Dynamics",
"author": "Humphrey W",
"doi-asserted-by": "crossref",
"first-page": "33",
"journal-title": "J. Mol. Graphics.",
"key": "ref67",
"unstructured": "Humphrey, W., Dalke, A., & Schulten, K. VMD - Visual Molecular Dynamics. J. Mol. Graphics. 14, 33–38 (1996).",
"volume": "14",
"year": "1996"
},
{
"DOI": "10.1002/jcc.20084",
"article-title": "UCSF Chimera - A visualization system for exploratory research and analysis",
"author": "Pettersen E",
"doi-asserted-by": "crossref",
"first-page": "1605",
"journal-title": "J. Comp. Chem.",
"key": "ref68",
"unstructured": "Pettersen, E. et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comp. Chem. 25, 1605–1612 (2004).",
"volume": "25",
"year": "2004"
},
{
"author": "Schrödinger Release 2022-4",
"key": "ref69",
"unstructured": "Schrödinger Release 2022-4: Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2022.",
"volume-title": "Protein Preparation Wizard; Epik, Schrödinger",
"year": "2022"
},
{
"DOI": "10.1002/jcc.20292",
"article-title": "Integrated modeling program applied chemical theory (IMPACT)",
"author": "Banks JL",
"doi-asserted-by": "crossref",
"first-page": "1752",
"journal-title": "J. Comp. Chem.",
"key": "ref70",
"unstructured": "Banks, J.L. et al. Integrated modeling program applied chemical theory (IMPACT). J. Comp. Chem. 26, 1752–1780 (2005).",
"volume": "26",
"year": "2005"
},
{
"key": "ref71",
"unstructured": "Schrödinger Release 2022-4: Glide, Schrödinger, LLC, New York, NY, 2022"
},
{
"author": "Schrödinger",
"key": "ref72",
"unstructured": "Schrödinger Release 2022-4: LigPrep, Schrödinger, LLC, New York, NY, 2022",
"year": "2022"
},
{
"DOI": "10.1002/prot.23106",
"article-title": "The VSGB 2.0 model: a next-generation energy model for high-resolution protein structure modeling",
"author": "Li J",
"doi-asserted-by": "crossref",
"first-page": "2794",
"journal-title": "Proteins.",
"key": "ref73",
"unstructured": "Li, J., Abel, R., Zhu, K., Cao, Y., Zhao, S., & Friesner RA. The VSGB 2.0 model: a next-generation energy model for high-resolution protein structure modeling. Proteins. 79, 2794–2812 (2011).",
"volume": "79",
"year": "2011"
},
{
"DOI": "10.1016/j.scitotenv.2013.06.081",
"article-title": "Validation of quantitative structure-activity relationship models to predict water-solubility of organic compounds",
"author": "Cappelli CI",
"doi-asserted-by": "crossref",
"first-page": "781",
"journal-title": "Sci. Total Environ",
"key": "ref74",
"unstructured": "Cappelli, C.I., Manganelli, S., Lombardo, A., Gissi, A., & Benfenati, E. Validation of quantitative structure-activity relationship models to predict water-solubility of organic compounds. Sci. Total Environ. 463–464, 781–789 (2013).",
"volume": "463–464",
"year": "2013"
},
{
"DOI": "10.1517/17460441.1.1.31",
"article-title": "In silico prediction of aqueous solubility",
"author": "Dearden JC",
"doi-asserted-by": "crossref",
"first-page": "31",
"journal-title": "Expet Opin. Drug Discov",
"key": "ref75",
"unstructured": "Dearden, J.C. In silico prediction of aqueous solubility. Expet Opin. Drug Discov. 1, 31–52 (2006).",
"volume": "1",
"year": "2006"
}
],
"reference-count": 75,
"references-count": 75,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.researchsquare.com/article/rs-3888947/v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach",
"type": "posted-content"
}