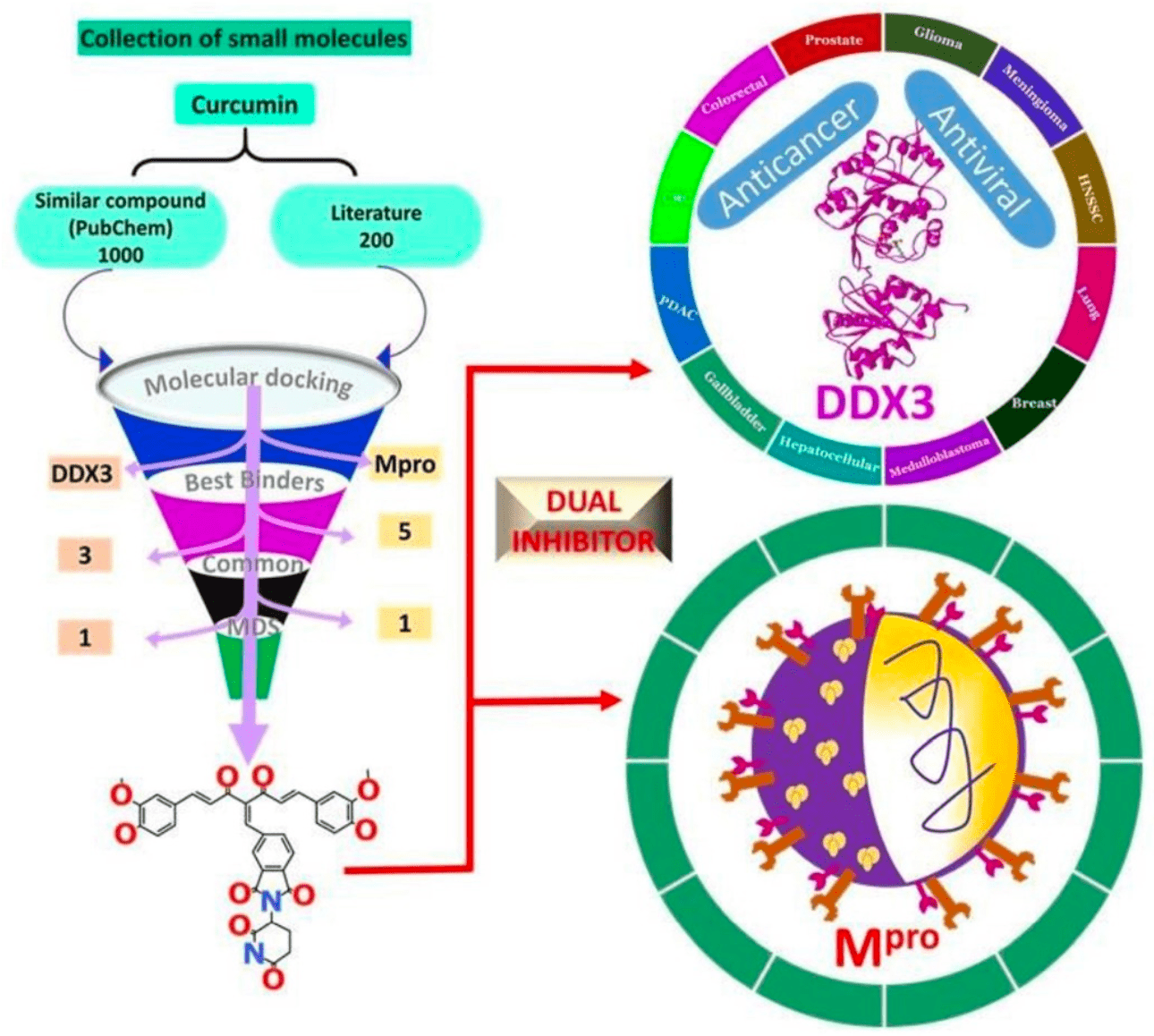
Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2
et al., International Journal of Molecular Sciences, doi:10.3390/ijms23031771, Feb 2022
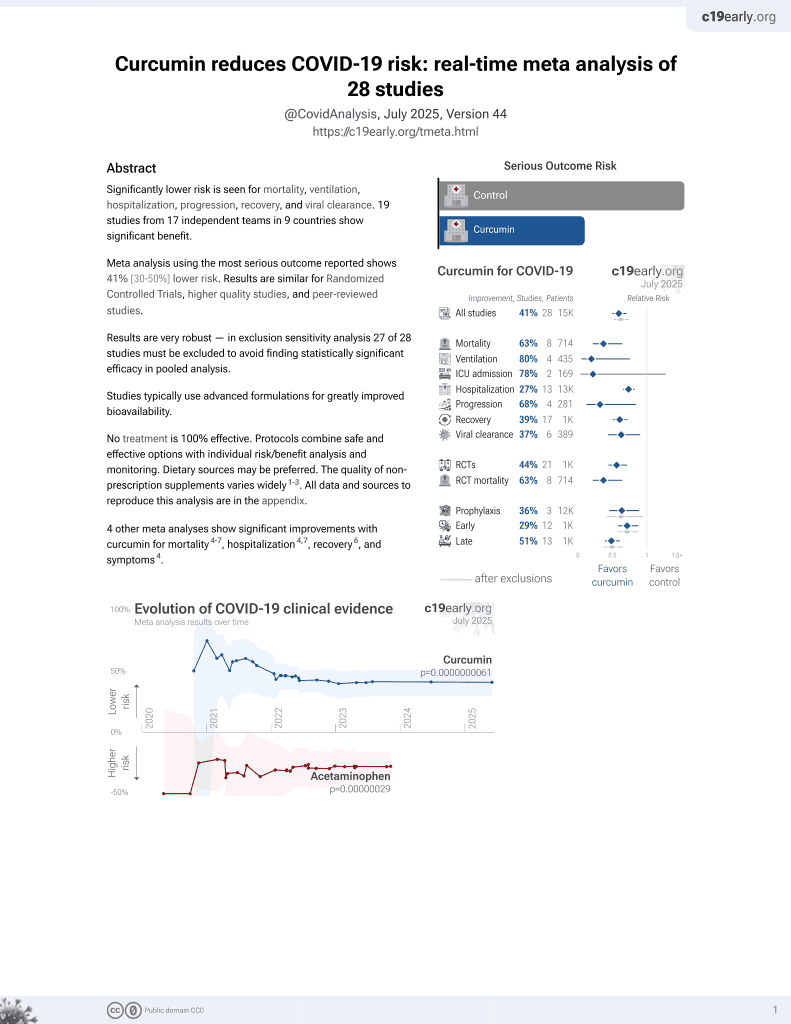
Curcumin for COVID-19
17th treatment shown to reduce risk in
February 2021, now with p = 0.0000000061 from 28 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico molecular dynamics simulation study finding a curcumin analogue (curA) as a promising dual inhibitor for SARS-CoV-2.
62 preclinical studies support the efficacy of curcumin for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with curcumin or metabolites via binding to the spikeA,1,5,6,11,16,18,24,27 (and specifically the receptor binding domainB,2,4,14,17,20 ), MproC,4-6,11,13,15-17,19,20,22,25,27,28,30,48 , RNA-dependent RNA polymeraseD,4-6,17,26 , PLproE,6, ACE2F,2,18,19,21 , nucleocapsidG,12,29 , nsp10H,29, and helicaseI,36 proteins, and inhibition of spike-ACE2 interactionJ,3.
In vitro studies demonstrate inhibition of the spikeA,41 (and specifically the receptor binding domainB,51), MproC,23,41,48,50 , ACE2F,51, and TMPRSS2K,51 proteins, and inhibition of spike-ACE2 interactionJ,3,34 .
In vitro studies demonstrate efficacy in Calu-3L,49, A549M,41, A549-ATN,31, 293TO,7, HEK293-hACE2P,23,39 , 293T/hACE2/TMPRSS2Q,40, Vero E6R,1,13,17,27,39,41,43,45,47,49 , and SH-SY5YS,38 cells.
Curcumin decreases pro-inflammatory cytokines induced by SARS-CoV-2 in peripheral blood mononuclear cells47, alleviates SARS-CoV-2 spike protein-induced mitochondrial membrane damage and oxidative stress7, may limit COVID-19 induced cardiac damage by inhibiting the NF-κB signaling pathway which mediates the profibrotic effects of the SARS-CoV-2 spike protein on cardiac fibroblasts35, is predicted to inhibit the interaction between the SARS-CoV-2 spike protein receptor binding domain and the human ACE2 receptor for the delta and omicron variants14, lowers ACE2 and STAT3, curbing lung inflammation and ARDS in preclinical COVID-19 models32, inhibits SARS-CoV-2 ORF3a ion channel activity, which contributes to viral pathogenicity and cytotoxicity42, has direct virucidal action by disrupting viral envelope integrity44, may inhibit viral replication and modulate inflammatory pathways like NF-κB via SIRT1 activation52, and can function as a photosensitizer in photodynamic therapy to generate reactive oxygen species that damage the virus44.
1.
Marzouk et al., Computational and Experimental Insights into the Antiviral Mechanism of Turmeric (Curcuma longa) against SARS-CoV-2 D614G, BIO Web of Conferences, doi:10.1051/bioconf/202519804002.
2.
Wu et al., Utilizing natural compounds as ligands to disrupt the binding of SARS-CoV-2 receptor-binding domain to angiotensin-converting enzyme 2, impeding viral infection, Phytochemistry Letters, doi:10.1016/j.phytol.2025.102999.
3.
Najimi et al., Phytochemical Inhibitors of SARS‐CoV‐2 Entry: Targeting the ACE2‐RBD Interaction with l‐Tartaric Acid, l‐Ascorbic Acid, and Curcuma longa Extract, ChemistrySelect, doi:10.1002/slct.202406035.
4.
Rajamanickam et al., Exploring the Potential of Siddha Formulation MilagaiKudineer-Derived Phytotherapeutics Against SARS-CoV-2: An In-Silico Investigation for Antiviral Intervention, Journal of Pharmacy and Pharmacology Research, doi:10.26502/fjppr.0105.
5.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
6.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
7.
Zhang et al., Computational Discovery of Mitochondrial Dysfunction Biomarkers in Severe SARS-CoV-2 Infection: Facilitating Pytomedicine Screening, Phytomedicine, doi:10.1016/j.phymed.2024.155784.
8.
Öztürkkan et al., In Silico investigation of the effects of curcuminoids on the spike protein of the omicron variant of SARS-CoV-2, Baku State University Journal of Chemistry and Material Sciences, 1:2, bsuj.bsu.edu.az/uploads/pdf/ec4204d62f7802de54e6092bf7860029.pdf.
9.
Yunze et al., Therapeutic effect and potential mechanism of curcumin, an active ingredient in Tongnao Decoction, on COVID-19 combined with stroke: a network pharmacology study and GEO database mining, Research Square, doi:10.21203/rs.3.rs-4329762/v1.
10.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
11.
Boseila et al., Throat spray formulated with virucidal Pharmaceutical excipients as an effective early prophylactic or treatment strategy against pharyngitis post-exposure to SARS CoV-2, European Journal of Pharmaceutics and Biopharmaceutics, doi:10.1016/j.ejpb.2024.114279.
12.
Hidayah et al., Bioinformatics study of curcumin, demethoxycurcumin, bisdemethoxycurcumin and cyclocurcumin compounds in Curcuma longa as an antiviral agent via nucleocapsid on SARS-CoV-2 inhibition, International Conference on Organic and Applied Chemistry, doi:10.1063/5.0197724.
13.
Singh et al., Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach, Research Square, doi:10.21203/rs.3.rs-3888947/v1.
14.
Kant et al., Structure-based drug discovery to identify SARS-CoV2 spike protein–ACE2 interaction inhibitors, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2300060.
15.
Naderi Beni et al., In silico studies of anti-oxidative and hot temperament-based phytochemicals as natural inhibitors of SARS-CoV-2 Mpro, PLOS ONE, doi:10.1371/journal.pone.0295014.
16.
Moschovou et al., Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms242115894.
17.
Eleraky et al., Curcumin Transferosome-Loaded Thermosensitive Intranasal in situ Gel as Prospective Antiviral Therapy for SARS-Cov-2, International Journal of Nanomedicine, doi:10.2147/IJN.S423251.
18.
Singh (B) et al., Computational studies to analyze effect of curcumin inhibition on coronavirus D614G mutated spike protein, The Seybold Report, doi:10.17605/OSF.IO/TKEXJ.
19.
Thapa et al., In-silico Approach for Predicting the Inhibitory Effect of Home Remedies on Severe Acute Respiratory Syndrome Coronavirus-2, Makara Journal of Science, doi:10.7454/mss.v27i3.1609.
20.
Srivastava et al., Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach, Stresses, doi:10.3390/stresses3030043.
21.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
22.
Winih Kinasih et al., Analisis in silico interaksi senyawa kurkuminoid terhadap enzim main protease 6LU7 dari SARS-CoV-2, Duta Pharma Journal, doi:10.47701/djp.v3i1.2904.
23.
Wu (B) et al., Potential Mechanism of Curcumin and Resveratrol against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-2780614/v1.
24.
Nag et al., Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2022.105552.
25.
Rampogu et al., Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2, International Journal of Molecular Sciences, doi:10.3390/ijms23031771.
26.
Singh (C) et al., Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2021.104965.
27.
Kandeil et al., Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2, Pathogens, doi:10.3390/pathogens10060758.
28.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
29.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
30.
Sekiou et al., In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2, ChemRxiv, doi:10.26434/chemrxiv.12181404.v1.
31.
Grüneberg et al., Dose-dependent antiviral effects of glycyrrhizin, curcumin, and harmaline against clinical SARS-CoV-2 isolates, including D614G, Omicron BA.5, and Omicron XBB.1, BMC Complementary Medicine and Therapies, doi:10.1186/s12906-026-05253-1.
32.
Aktay et al., Oral Administration of Water-Soluble Curcumin Complex Prevents ARDS With the Potential for COVID-19 Treatment, Phytotherapy Research, doi:10.1002/ptr.70046.
33.
Olubiyi et al., Novel dietary herbal preparations with inhibitory activities against multiple SARS-CoV-2 targets: A multidisciplinary investigation into antiviral activities, Food Chemistry Advances, doi:10.1016/j.focha.2025.100969.
34.
Emam et al., Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors, AMB Express, doi:10.1186/s13568-024-01739-8.
35.
Van Tin et al., Spike Protein of SARS-CoV-2 Activates Cardiac Fibrogenesis through NLRP3 Inflammasomes and NF-κB Signaling, Cells, doi:10.3390/cells13161331.
36.
Li et al., Thermal shift assay (TSA)-based drug screening strategy for rapid discovery of inhibitors against the Nsp13 helicase of SARS-CoV-2, Animals and Zoonoses, doi:10.1016/j.azn.2024.06.001.
37.
Kamble et al., Nanoparticulate curcumin spray imparts prophylactic and therapeutic properties against SARS-CoV-2, Emergent Materials, doi:10.1007/s42247-024-00754-6.
38.
Nicoliche et al., Antiviral, anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-61662-7.
39.
Nittayananta et al., A novel film spray containing curcumin inhibits SARS-CoV-2 and influenza virus infection and enhances mucosal immunity, Virology Journal, doi:10.1186/s12985-023-02282-x.
40.
Septisetyani et al., Curcumin and turmeric extract inhibited SARS-CoV-2 pseudovirus cell entry and Spike mediated cell fusion, bioRxiv, doi:10.1101/2023.09.28.560070.
41.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
42.
Fam et al., Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Scientific Reports, doi:10.1038/s41598-023-31764-9.
43.
Teshima et al., Antiviral activity of curcumin and its analogs selected by an artificial intelligence-supported activity prediction system in SARS-CoV-2-infected VeroE6 cells, Natural Product Research, doi:10.1080/14786419.2023.2194647.
44.
Zupin et al., Optimization of Anti-SARS-CoV-2 Treatments Based on Curcumin, Used Alone or Employed as a Photosensitizer, Viruses, doi:10.3390/v14102132.
45.
Leka et al., In vitro antiviral activity against SARS-CoV-2 of common herbal medicinal extracts and their bioactive compounds, Phytotherapy Research, doi:10.1002/ptr.7463.
46.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
47.
Marín-Palma et al., Curcumin Inhibits In Vitro SARS-CoV-2 Infection In Vero E6 Cells through Multiple Antiviral Mechanisms, Molecules, doi:10.3390/molecules26226900.
48.
Bahun et al., Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols, Food Chemistry, doi:10.1016/j.foodchem.2021.131594.
49.
Bormann et al., Turmeric Root and Its Bioactive Ingredient Curcumin Effectively Neutralize SARS-CoV-2 In Vitro, Viruses, doi:10.3390/v13101914.
50.
Guijarro-Real et al., Potential In Vitro Inhibition of Selected Plant Extracts against SARS-CoV-2 Chymotripsin-Like Protease (3CLPro) Activity, Foods, doi:10.3390/foods10071503.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
g.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
h.
Non-structural protein 10 (nsp10) serves as an RNA chaperone and stabilizes conformations of nsp12 and nsp14 in the replicase-transcriptase complex, which synthesizes new viral RNAs. Nsp10 disruption may destabilize replicase-transcriptase complex activity.
i.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
j.
The interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor is a primary method of viral entry, inhibiting this interaction can prevent the virus from attaching to and entering host cells, halting infection at an early stage.
k.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
l.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
m.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
n.
A549-AT is a human lung carcinoma cell line stably transfected with ACE2 and TMPRSS2 receptors. Unlike the parental line, this overexpression ensures stable infection and enhanced viral entry, allowing for the evaluation of antiviral efficacy against various SARS-CoV-2 variants.
o.
293T is a human embryonic kidney cell line that can be engineered for high ACE2 expression and SARS-CoV-2 susceptibility. 293T cells are easily transfected and support high protein expression.
p.
HEK293-hACE2 is a human embryonic kidney cell line with high ACE2 expression and SARS-CoV-2 susceptibility. Cells have been transfected with a plasmid to express the human ACE2 (hACE2) protein.
q.
293T/hACE2/TMPRSS2 is a human embryonic kidney cell line engineered for high ACE2 and TMPRSS2 expression, which mimics key aspects of human infection. 293T/hACE2/TMPRSS2 cells are very susceptible to SARS-CoV-2 infection.
r.
Vero E6 is an African green monkey kidney cell line with low/no ACE2 expression and high SARS-CoV-2 susceptibility. The cell line is easy to maintain and supports robust viral replication, however the monkey origin may not accurately represent human responses.
s.
SH-SY5Y is a human neuroblastoma cell line that exhibits neuronal phenotypes. It is commonly used as an in vitro model for studying neurotoxicity, neurodegenerative diseases, and neuronal differentiation.
Rampogu et al., 4 Feb 2022, peer-reviewed, 5 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2
International Journal of Molecular Sciences, doi:10.3390/ijms23031771
Recently, the world has been witnessing a global pandemic with no effective therapeutics yet, while cancer continues to be a major disease claiming many lives. The natural compound curcumin is bestowed with multiple medicinal applications in addition to demonstrating antiviral and anticancer activities. In order to elucidate the impact of curcumin on COVID-19 and cancer, the current investigation has adapted several computational techniques to unfold its possible inhibitory activity. Accordingly, curcumin and similar compounds and analogues were retrieved and assessed for their binding affinities at the binding pocket of SARS-CoV-2 main protease and DDX3. The best binding pose was escalated to molecular dynamics simulation (MDS) studies to assess the time dependent stability. Our findings have rendered one compound that has demonstrated good molecular dock score complemented by key residue interactions and have shown stable MDS results inferred by root mean square deviation (RMSD), radius of gyration (Rg), binding mode, hydrogen bond interactions, and interaction energy. Essential dynamics results have shown that the systemadapts minimum energy conformation to attain a stable state. The discovered compound (curA) could act as plausible inhibitor against SARS-CoV-2 and DDX3. Furthermore, curA could serve as a chemical scaffold for designing and developing new compounds.
Supplementary Materials: The following supporting information can be downloaded at: https: //www.mdpi.com/article/10.3390/ijms23031771/s1.
Conflicts of Interest: The authors declare no conflict of interest.
References
Aier, Varadwaj, Raj, Structural insights into conformational stability of both wild-type and mutant EZH2 receptor, Sci. Rep, doi:10.1038/srep34984
Alexpandi, De Mesquita, Pandian, Ravi, Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Analysis, Front. Microbiol, doi:10.3389/fmicb.2020.01796
Atanasov, Zotchev, Dirsch, Orhan, Banach et al., Natural products in drug discovery: Advances and opportunities, Nat. Rev. Drug Discov, doi:10.1038/s41573-020-00114-z
Bhardwaj, Singh, Das, Purohit, Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs, Comput. Biol. Med, doi:10.1016/j.compbiomed.2020.104117
Bimonte, Barbieri, Palma, Rea, Luciano et al., Dissecting the Role of Curcumin in Tumour Growth and Angiogenesis in Mouse Model of Human Breast Cancer, Biomed Res. Int, doi:10.1155/2015/878134
Bol, Xie, Raman, DDX3, a potential target for cancer treatment, Mol. Cancer, doi:10.1186/s12943-015-0461-7
Botlagunta, Kollapalli, Kakarla, Gajarla, Gade et al., In vitro anti-cancer activity of doxorubicin against human RNA helicase, DDX3, Bioinformation, doi:10.6026/97320630012347
Ciccosanti, Di Rienzo, Romagnoli, Colavita, Refolo et al., Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection, Antivir. Res, doi:10.1016/j.antiviral.2021.105064
Fakhar, Khan, Alomar, Alkhuriji, Ahmad, ABBV-744 as a potential inhibitor of SARS-CoV-2 main protease enzyme against COVID-19, Sci. Rep, doi:10.1038/s41598-020-79918-3
Frecer, Miertus, Antiviral agents against COVID-19: Structure-based design of specific peptidomimetic inhibitors of SARS-CoV-2 main protease, RSC Adv, doi:10.1039/D0RA08304F
Gahlawat, Kumar, Kumar, Sandhu, Singh et al., Structure-Based Virtual Screening to Discover Potential Lead Molecules for the SARS-CoV-2 Main Protease, J. Chem. Inf. Model, doi:10.1021/acs.jcim.0c00546
Ghahremanpour, Tirado-Rives, Deshmukh, Ippolito, Zhang et al., Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2, ACS Med. Chem. Lett, doi:10.1021/acsmedchemlett.0c00521
Giordano, Tommonaro, Curcumin and Cancer, Nutrients, doi:10.3390/nu11102376
Guo, Xie, Lei, Liu, Zhang et al., Discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 via virtual screening and biochemical evaluation, Bioorg. Chem, doi:10.1016/j.bioorg.2021.104767
He, Zhang, Yang, Wang, Zhao et al., A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review), Oncol. Rep, doi:10.3892/or.2018.6203
Hospital, Goñi, Orozco, Gelpí, Molecular dynamics simulations: Advances and applications, Adv. Appl. Bioinform. Chem, doi:10.2147/AABC.S70333
Hu, Xu, Meng, Huang, Sun, Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells, Exp. Ther. Med, doi:10.3892/etm.2018.6345
Humphrey, Dalke, Schulten, Vmd, Visual molecular dynamics, J. Mol. Graph, doi:10.1016/0263-7855(96)00018-5
Högbom, Collins, Van Den Berg, Jenvert, Karlberg et al., Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP, J. Mol. Biol, doi:10.1016/j.jmb.2007.06.050
Jena, Kanungo, Nayak, Chainy, Dandapat, Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: Insights from computational studies, Sci. Rep, doi:10.1038/s41598-021-81462-7
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kim, Chen, Cheng, Gindulyte, He et al., PubChem in 2021: New data content and improved web interfaces, Nucleic Acids Res, doi:10.1093/nar/gkaa971
Kumar, Bhardwaj, Kumar, Gehi, Kapuganti et al., Reprofiling of approved drugs against SARS-CoV-2 main protease: An in-silico study, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1845976
Kumar, Nyodu, Maurya, Saxena, Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Epidemiol. Pathog. Diagn. Ther
Kumar, Sodhi, Singh, Addressing the potential role of curcumin in the prevention of COVID-19 by targeting the Nsp9 replicase protein through molecular docking, Arch. Microbiol, doi:10.1007/s00203-020-02163-9
Kumar, Swetha, Anbarasu, Ramaiah, Computational analysis reveals the association of threonine 118 methionine mutation in PMP22 resulting in CMT-1A, Adv. Bioinform, doi:10.1155/2014/502618
Li, Li, Huang, Wu, Liu et al., Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs, BioRxiv, doi:10.1073/pnas.2010470117
Lin, Hsu, Lin, Antiviral natural products and herbal medicines, J. Tradit. Complement. Med
Liu, Ying, The Inhibitory Effect of Curcumin on Virus-Induced Cytokine Storm and Its Potential Use in the Associated Severe Pneumonia, Front. Cell Dev. Biol, doi:10.3389/fcell.2020.00479
Manoharan, Haridas, Vasanthakumar, Muthu, Thavoorullah et al., Curcumin: A Wonder Drug as a Preventive Measure for COVID19 Management, Indian J. Clin. Biochem, doi:10.1007/s12291-020-00902-9
Mansouri, Rasoulpoor, Daneshkhah, Abolfathi, Salari et al., Clinical effects of curcumin in enhancing cancer therapy: A systematic review, BMC Cancer, doi:10.1186/s12885-020-07256-8
Marín-Palma, Tabares-Guevara, Zapata-Cardona, Flórez-Álvarez, Yepes et al., Curcumin Inhibits in Vitro SARS-CoV-2 Infection in Vero E6 Cells through Multiple Antiviral Mechanisms, Molecules, doi:10.3390/molecules26226900
Mathew, Hsu, Antiviral potential of curcumin, J. Funct. Foods, doi:10.1016/j.jff.2017.12.017
Mishra, Pandey, Sharma, Malik, Mongre et al., Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: An integrated computational approach, Brief. Bioinform, doi:10.1093/bib/bbaa378
Mo, Liang, Su, Li, Chen et al., DDX3X: Structure, physiologic functions and cancer, Mol. Cancer, doi:10.1186/s12943-021-01325-7
Mohammed, Rashid-Doubell, Taha, Cassidy, Fredericks, Effects of curcumin complexes on MDA-MB-231 breast cancer cell proliferation, Int. J. Oncol, doi:10.3892/ijo.2020.5065
Naqvi, Fatima, Mohammad, Fatima, Singh et al., Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach, Biochim. Biophys. Acta Mol. Basis Dis, doi:10.1016/j.bbadis.2020.165878
Nguyen, Gao, Chen, Wang, Wei, Unveiling the molecular mechanism of SARS-CoV-2 main protease inhibition from 137 crystal structures using algebraic topology and deep learning, Chem. Sci, doi:10.1039/D0SC04641H
Noureddin, El-Shishtawy, Al-Footy, Curcumin analogues and their hybrid molecules as multifunctional drugs, Eur. J. Med. Chem, doi:10.1016/j.ejmech.2019.111631
Patel, Rajendran, Shah, Patel, Pakala et al., Virtual screening of curcumin and its analogs against the spike surface glycoprotein of SARS-CoV-2 and SARS-CoV, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1868338
Quaranta, Lottini, Chesi, Contrafatto, Russotto et al., DDX3 inhibitors show antiviral activity against positive-sense single-stranded RNA viruses but not against negative-sense single-stranded RNA viruses: The coxsackie B model, Antivir. Res, doi:10.1016/j.antiviral.2020.104750
Rampogu, Kim, Son, Baek, Park et al., A computational approach with biological evaluation: Combinatorial treatment of curcumin and exemestane synergistically regulates ddx3 expression in cancer cell lines, Biomolecules, doi:10.3390/biom10060857
Rampogu, Lee, Old Drugs for New Purpose-Fast Pace Therapeutic Identification for SARS-CoV-2 Infections by Pharmacophore Guided Drug Repositioning Approach, Bull. Korean Chem. Soc, doi:10.1002/bkcs.12171
Rampogu, Lemuel, Lee, Virtual screening, molecular docking, molecular dynamics simulations and free energy calculations to discover potential DDX3 inhibitors, Adv. Cancer Biol.-Metastasis, doi:10.1016/j.adcanc.2021.100022
Rampogu, Parameswaran, Lemuel, Lee, Exploring the Therapeutic Ability of Fenugreek against Type 2 Diabetes and Breast Cancer Employing Molecular Docking and Molecular Dynamics Simulations, Evid.-Based Complement. Altern. Med, doi:10.1155/2018/1943203
Sacco, Ma, Lagarias, Gao, Townsend et al., Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and Cathepsin L, Sci. Adv, doi:10.1126/sciadv.abe0751
Samal, Routray, Veeramachaneni, Dash, Botlagunta, Ketorolac salt is a newly discovered DDX3 inhibitor to treat oral cancer, Sci. Rep, doi:10.1038/srep09982
Shanmugarajan, Prabitha, Kumar, Suresh, Curcumin to inhibit binding of spike glycoprotein to ACE2 receptors: Computational modelling, simulations, and ADMET studies to explore curcuminoids against novel SARS-CoV-2 targets, RSC Adv, doi:10.1039/D0RA03167D
Soni, Mehta, Ratre, Tiwari, Amit et al., Curcumin, a traditional spice component, can hold the promise against COVID-19?, Eur. J. Pharmacol, doi:10.1016/j.ejphar.2020.173551
Thimmulappa, Mudnakudu-Nagaraju, Shivamallu, Subramaniam, Radhakrishnan et al., Antiviral and immunomodulatory activity of curcumin: A case for prophylactic therapy for COVID-19, Heliyon, doi:10.1016/j.heliyon.2021.e06350
Thulasi Raman, Liu, Pyo, Cui, Xu et al., DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation, J. Virol, doi:10.1128/JVI.03010-15
Valiente-Echeverría, Hermoso, Soto-Rifo, RNA helicase DDX3: At the crossroad of viral replication and antiviral immunity, Rev. Med. Virol, doi:10.1002/rmv.1845
Van Der Spoel, Lindahl, Hess, Groenhof, Mark et al., GROMACS: Fast, flexible, and free, J. Comput. Chem, doi:10.1002/jcc.20291
Van Voss, Vesuna, Trumpi, Brilliant, Berlinicke et al., Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer, Oncotarget, doi:10.18632/oncotarget.4873
Vyas, Dandawate, Padhye, Ahmad, Sarkar, Perspectives on new synthetic curcumin analogs and their potential anticancer properties, Curr. Pharm. Des
Wu, Robertson, Brooks, Vieth, Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm, J. Comput. Chem, doi:10.1002/jcc.10306
Wu, Wu, Liu, Yang, The SARS-CoV-2 outbreak: What we know, Int. J. Infect. Dis, doi:10.1016/j.ijid.2020.03.004
Yang, Xiao, Ye, He, Sun et al., SARS-CoV-2: Characteristics and current advances in research, Virol. J, doi:10.1186/s12985-020-01369-z
Zoete, Cuendet, Grosdidier, Michielin, SwissParam: A fast force field generation tool for smallorganic molecules, J. Comput. Chem, doi:10.1002/jcc.21816
DOI record:
{
"DOI": "10.3390/ijms23031771",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms23031771",
"abstract": "<jats:p>Recently, the world has been witnessing a global pandemic with no effective therapeutics yet, while cancer continues to be a major disease claiming many lives. The natural compound curcumin is bestowed with multiple medicinal applications in addition to demonstrating antiviral and anticancer activities. In order to elucidate the impact of curcumin on COVID-19 and cancer, the current investigation has adapted several computational techniques to unfold its possible inhibitory activity. Accordingly, curcumin and similar compounds and analogues were retrieved and assessed for their binding affinities at the binding pocket of SARS-CoV-2 main protease and DDX3. The best binding pose was escalated to molecular dynamics simulation (MDS) studies to assess the time dependent stability. Our findings have rendered one compound that has demonstrated good molecular dock score complemented by key residue interactions and have shown stable MDS results inferred by root mean square deviation (RMSD), radius of gyration (Rg), binding mode, hydrogen bond interactions, and interaction energy. Essential dynamics results have shown that the systemadapts minimum energy conformation to attain a stable state. The discovered compound (curA) could act as plausible inhibitor against SARS-CoV-2 and DDX3. Furthermore, curA could serve as a chemical scaffold for designing and developing new compounds.</jats:p>",
"alternative-id": [
"ijms23031771"
],
"author": [
{
"affiliation": [],
"family": "Rampogu",
"given": "Shailima",
"sequence": "first"
},
{
"affiliation": [],
"family": "Lee",
"given": "Gihwan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Park",
"given": "Jun Sung",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lee",
"given": "Keun Woo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kim",
"given": "Myeong Ok",
"sequence": "additional"
}
],
"container-title": [
"International Journal of Molecular Sciences"
],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
2,
4
]
],
"date-time": "2022-02-04T16:35:17Z",
"timestamp": 1643992517000
},
"deposited": {
"date-parts": [
[
2022,
2,
4
]
],
"date-time": "2022-02-04T17:00:46Z",
"timestamp": 1643994046000
},
"funder": [
{
"award": [
"NRF-2018M3A9A7057263",
"2020M3E5D9080660"
],
"name": "This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government"
}
],
"indexed": {
"date-parts": [
[
2022,
4,
1
]
],
"date-time": "2022-04-01T16:18:55Z",
"timestamp": 1648829935055
},
"is-referenced-by-count": 0,
"issn-type": [
{
"type": "electronic",
"value": "1422-0067"
}
],
"issue": "3",
"issued": {
"date-parts": [
[
2022,
2,
4
]
]
},
"journal-issue": {
"issue": "3",
"published-online": {
"date-parts": [
[
2022,
2
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2022,
2,
4
]
],
"date-time": "2022-02-04T00:00:00Z",
"timestamp": 1643932800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/23/3/1771/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1771",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2022,
2,
4
]
]
},
"published-online": {
"date-parts": [
[
2022,
2,
4
]
]
},
"publisher": "MDPI AG",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/23/3/1771"
}
},
"score": 1,
"short-container-title": [
"IJMS"
],
"short-title": [],
"source": "Crossref",
"subject": [
"Inorganic Chemistry",
"Organic Chemistry",
"Physical and Theoretical Chemistry",
"Computer Science Applications",
"Spectroscopy",
"Molecular Biology",
"General Medicine",
"Catalysis"
],
"subtitle": [],
"title": [
"Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2"
],
"type": "journal-article",
"volume": "23"
}