Structure-Based Design and In-Silico Evaluation of Computationally Proposed Curcumin Derivatives as Potential Inhibitors of the Coronaviral PLpro Enzymes
, H., Pharmaceuticals, doi:10.3390/ph18060798, May 2025
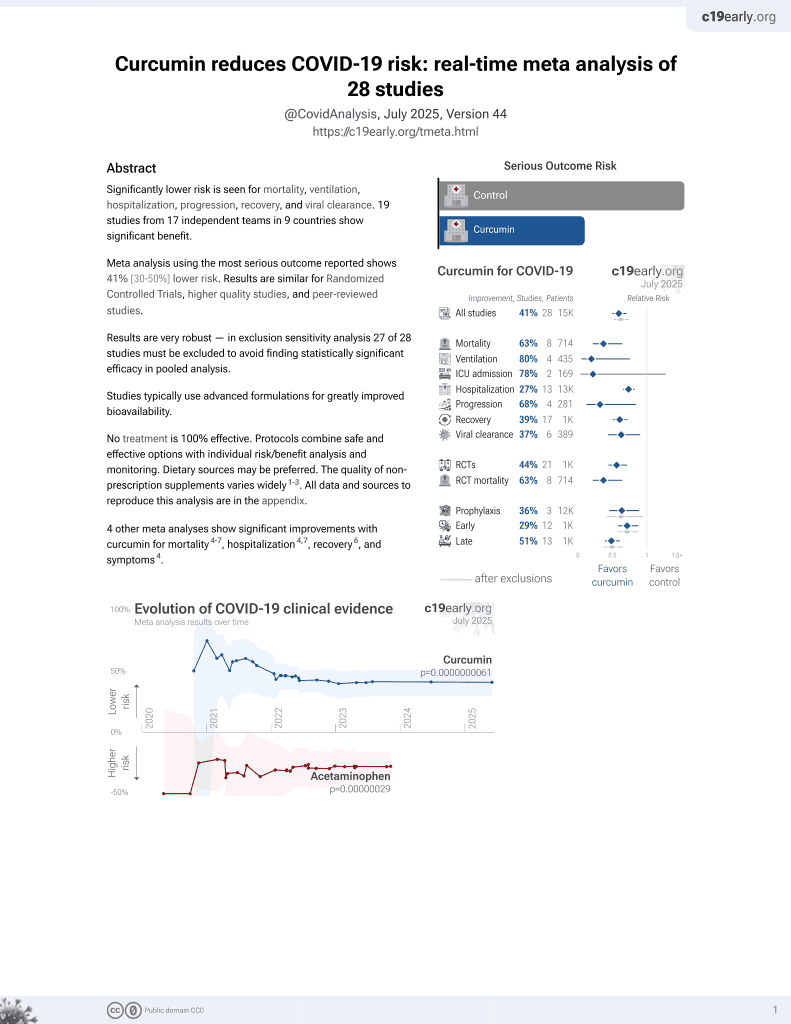
Curcumin for COVID-19
17th treatment shown to reduce risk in
February 2021, now with p = 0.0000000061 from 28 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
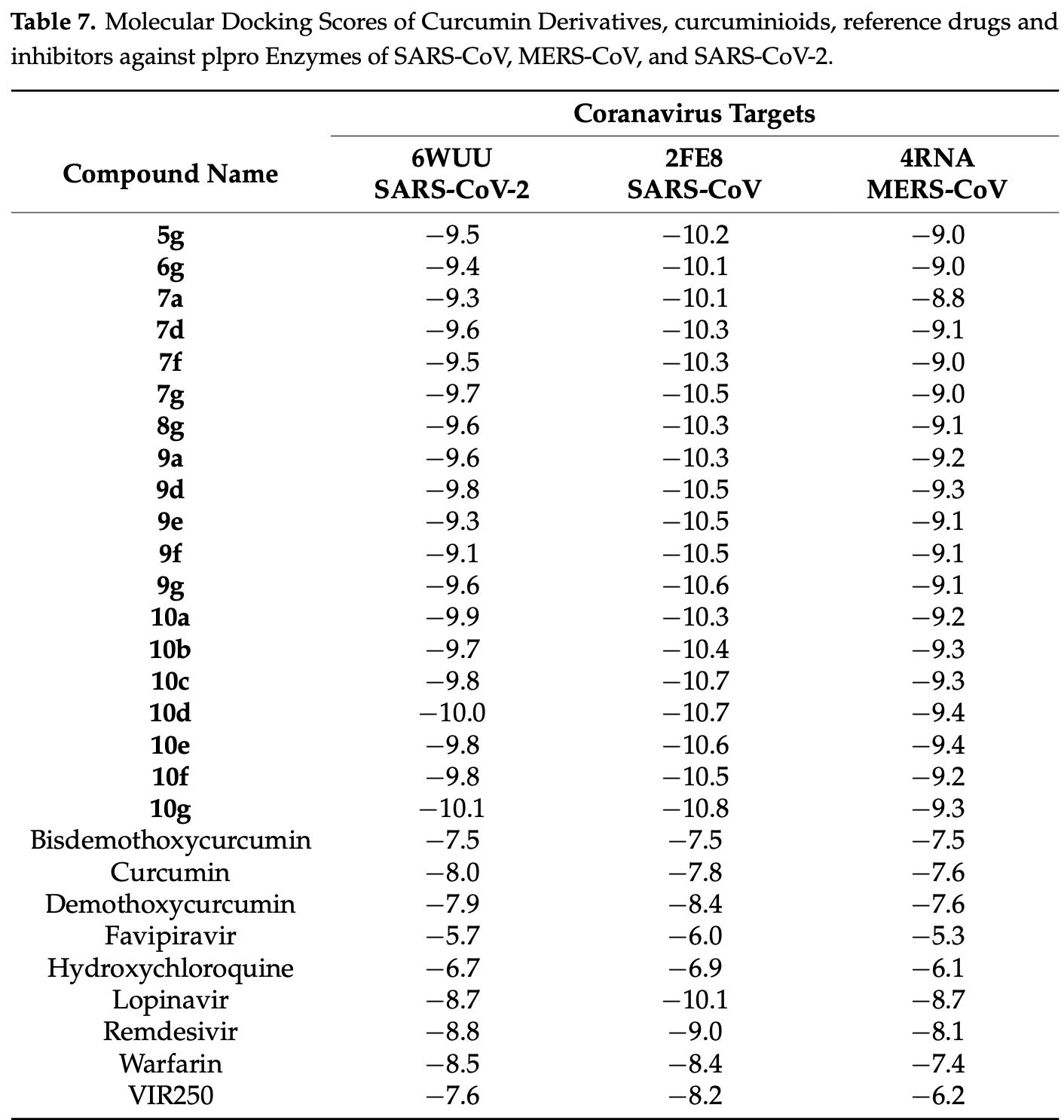
In silico study showing that novel curcumin derivatives strongly bind to papain-like protease (PLpro) enzymes from SARS-CoV, MERS-CoV, and SARS-CoV-2 with superior binding affinities compared to natural curcumin and reference antivirals.
62 preclinical studies support the efficacy of curcumin for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with curcumin or metabolites via binding to the spikeA,1,5,6,11,16,18,24,27 (and specifically the receptor binding domainB,2,4,14,17,20 ), MproC,4-6,11,13,15-17,19,20,22,25,27,28,30,48 , RNA-dependent RNA polymeraseD,4-6,17,26 , PLproE,6, ACE2F,2,18,19,21 , nucleocapsidG,12,29 , nsp10H,29, and helicaseI,36 proteins, and inhibition of spike-ACE2 interactionJ,3.
In vitro studies demonstrate inhibition of the spikeA,41 (and specifically the receptor binding domainB,51), MproC,23,41,48,50 , ACE2F,51, and TMPRSS2K,51 proteins, and inhibition of spike-ACE2 interactionJ,3,34 .
In vitro studies demonstrate efficacy in Calu-3L,49, A549M,41, A549-ATN,31, 293TO,7, HEK293-hACE2P,23,39 , 293T/hACE2/TMPRSS2Q,40, Vero E6R,1,13,17,27,39,41,43,45,47,49 , and SH-SY5YS,38 cells.
Curcumin decreases pro-inflammatory cytokines induced by SARS-CoV-2 in peripheral blood mononuclear cells47, alleviates SARS-CoV-2 spike protein-induced mitochondrial membrane damage and oxidative stress7, may limit COVID-19 induced cardiac damage by inhibiting the NF-κB signaling pathway which mediates the profibrotic effects of the SARS-CoV-2 spike protein on cardiac fibroblasts35, is predicted to inhibit the interaction between the SARS-CoV-2 spike protein receptor binding domain and the human ACE2 receptor for the delta and omicron variants14, lowers ACE2 and STAT3, curbing lung inflammation and ARDS in preclinical COVID-19 models32, inhibits SARS-CoV-2 ORF3a ion channel activity, which contributes to viral pathogenicity and cytotoxicity42, has direct virucidal action by disrupting viral envelope integrity44, may inhibit viral replication and modulate inflammatory pathways like NF-κB via SIRT1 activation52, and can function as a photosensitizer in photodynamic therapy to generate reactive oxygen species that damage the virus44.
1.
Marzouk et al., Computational and Experimental Insights into the Antiviral Mechanism of Turmeric (Curcuma longa) against SARS-CoV-2 D614G, BIO Web of Conferences, doi:10.1051/bioconf/202519804002.
2.
Wu et al., Utilizing natural compounds as ligands to disrupt the binding of SARS-CoV-2 receptor-binding domain to angiotensin-converting enzyme 2, impeding viral infection, Phytochemistry Letters, doi:10.1016/j.phytol.2025.102999.
3.
Najimi et al., Phytochemical Inhibitors of SARS‐CoV‐2 Entry: Targeting the ACE2‐RBD Interaction with l‐Tartaric Acid, l‐Ascorbic Acid, and Curcuma longa Extract, ChemistrySelect, doi:10.1002/slct.202406035.
4.
Rajamanickam et al., Exploring the Potential of Siddha Formulation MilagaiKudineer-Derived Phytotherapeutics Against SARS-CoV-2: An In-Silico Investigation for Antiviral Intervention, Journal of Pharmacy and Pharmacology Research, doi:10.26502/fjppr.0105.
5.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
6.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
7.
Zhang et al., Computational Discovery of Mitochondrial Dysfunction Biomarkers in Severe SARS-CoV-2 Infection: Facilitating Pytomedicine Screening, Phytomedicine, doi:10.1016/j.phymed.2024.155784.
8.
Öztürkkan et al., In Silico investigation of the effects of curcuminoids on the spike protein of the omicron variant of SARS-CoV-2, Baku State University Journal of Chemistry and Material Sciences, 1:2, bsuj.bsu.edu.az/uploads/pdf/ec4204d62f7802de54e6092bf7860029.pdf.
9.
Yunze et al., Therapeutic effect and potential mechanism of curcumin, an active ingredient in Tongnao Decoction, on COVID-19 combined with stroke: a network pharmacology study and GEO database mining, Research Square, doi:10.21203/rs.3.rs-4329762/v1.
10.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
11.
Boseila et al., Throat spray formulated with virucidal Pharmaceutical excipients as an effective early prophylactic or treatment strategy against pharyngitis post-exposure to SARS CoV-2, European Journal of Pharmaceutics and Biopharmaceutics, doi:10.1016/j.ejpb.2024.114279.
12.
Hidayah et al., Bioinformatics study of curcumin, demethoxycurcumin, bisdemethoxycurcumin and cyclocurcumin compounds in Curcuma longa as an antiviral agent via nucleocapsid on SARS-CoV-2 inhibition, International Conference on Organic and Applied Chemistry, doi:10.1063/5.0197724.
13.
Singh et al., Unlocking the potential of phytochemicals in inhibiting SARS-CoV-2 M Pro protein - An in-silico and cell-based approach, Research Square, doi:10.21203/rs.3.rs-3888947/v1.
14.
Kant et al., Structure-based drug discovery to identify SARS-CoV2 spike protein–ACE2 interaction inhibitors, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2300060.
15.
Naderi Beni et al., In silico studies of anti-oxidative and hot temperament-based phytochemicals as natural inhibitors of SARS-CoV-2 Mpro, PLOS ONE, doi:10.1371/journal.pone.0295014.
16.
Moschovou et al., Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms242115894.
17.
Eleraky et al., Curcumin Transferosome-Loaded Thermosensitive Intranasal in situ Gel as Prospective Antiviral Therapy for SARS-Cov-2, International Journal of Nanomedicine, doi:10.2147/IJN.S423251.
18.
Singh (B) et al., Computational studies to analyze effect of curcumin inhibition on coronavirus D614G mutated spike protein, The Seybold Report, doi:10.17605/OSF.IO/TKEXJ.
19.
Thapa et al., In-silico Approach for Predicting the Inhibitory Effect of Home Remedies on Severe Acute Respiratory Syndrome Coronavirus-2, Makara Journal of Science, doi:10.7454/mss.v27i3.1609.
20.
Srivastava et al., Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach, Stresses, doi:10.3390/stresses3030043.
21.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
22.
Winih Kinasih et al., Analisis in silico interaksi senyawa kurkuminoid terhadap enzim main protease 6LU7 dari SARS-CoV-2, Duta Pharma Journal, doi:10.47701/djp.v3i1.2904.
23.
Wu (B) et al., Potential Mechanism of Curcumin and Resveratrol against SARS-CoV-2, Research Square, doi:10.21203/rs.3.rs-2780614/v1.
24.
Nag et al., Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2022.105552.
25.
Rampogu et al., Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2, International Journal of Molecular Sciences, doi:10.3390/ijms23031771.
26.
Singh (C) et al., Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2021.104965.
27.
Kandeil et al., Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2, Pathogens, doi:10.3390/pathogens10060758.
28.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
29.
Suravajhala et al., Comparative Docking Studies on Curcumin with COVID-19 Proteins, Preprints, doi:10.20944/preprints202005.0439.v3.
30.
Sekiou et al., In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2, ChemRxiv, doi:10.26434/chemrxiv.12181404.v1.
31.
Grüneberg et al., Dose-dependent antiviral effects of glycyrrhizin, curcumin, and harmaline against clinical SARS-CoV-2 isolates, including D614G, Omicron BA.5, and Omicron XBB.1, BMC Complementary Medicine and Therapies, doi:10.1186/s12906-026-05253-1.
32.
Aktay et al., Oral Administration of Water-Soluble Curcumin Complex Prevents ARDS With the Potential for COVID-19 Treatment, Phytotherapy Research, doi:10.1002/ptr.70046.
33.
Olubiyi et al., Novel dietary herbal preparations with inhibitory activities against multiple SARS-CoV-2 targets: A multidisciplinary investigation into antiviral activities, Food Chemistry Advances, doi:10.1016/j.focha.2025.100969.
34.
Emam et al., Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors, AMB Express, doi:10.1186/s13568-024-01739-8.
35.
Van Tin et al., Spike Protein of SARS-CoV-2 Activates Cardiac Fibrogenesis through NLRP3 Inflammasomes and NF-κB Signaling, Cells, doi:10.3390/cells13161331.
36.
Li et al., Thermal shift assay (TSA)-based drug screening strategy for rapid discovery of inhibitors against the Nsp13 helicase of SARS-CoV-2, Animals and Zoonoses, doi:10.1016/j.azn.2024.06.001.
37.
Kamble et al., Nanoparticulate curcumin spray imparts prophylactic and therapeutic properties against SARS-CoV-2, Emergent Materials, doi:10.1007/s42247-024-00754-6.
38.
Nicoliche et al., Antiviral, anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-61662-7.
39.
Nittayananta et al., A novel film spray containing curcumin inhibits SARS-CoV-2 and influenza virus infection and enhances mucosal immunity, Virology Journal, doi:10.1186/s12985-023-02282-x.
40.
Septisetyani et al., Curcumin and turmeric extract inhibited SARS-CoV-2 pseudovirus cell entry and Spike mediated cell fusion, bioRxiv, doi:10.1101/2023.09.28.560070.
41.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
42.
Fam et al., Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Scientific Reports, doi:10.1038/s41598-023-31764-9.
43.
Teshima et al., Antiviral activity of curcumin and its analogs selected by an artificial intelligence-supported activity prediction system in SARS-CoV-2-infected VeroE6 cells, Natural Product Research, doi:10.1080/14786419.2023.2194647.
44.
Zupin et al., Optimization of Anti-SARS-CoV-2 Treatments Based on Curcumin, Used Alone or Employed as a Photosensitizer, Viruses, doi:10.3390/v14102132.
45.
Leka et al., In vitro antiviral activity against SARS-CoV-2 of common herbal medicinal extracts and their bioactive compounds, Phytotherapy Research, doi:10.1002/ptr.7463.
46.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
47.
Marín-Palma et al., Curcumin Inhibits In Vitro SARS-CoV-2 Infection In Vero E6 Cells through Multiple Antiviral Mechanisms, Molecules, doi:10.3390/molecules26226900.
48.
Bahun et al., Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols, Food Chemistry, doi:10.1016/j.foodchem.2021.131594.
49.
Bormann et al., Turmeric Root and Its Bioactive Ingredient Curcumin Effectively Neutralize SARS-CoV-2 In Vitro, Viruses, doi:10.3390/v13101914.
50.
Guijarro-Real et al., Potential In Vitro Inhibition of Selected Plant Extracts against SARS-CoV-2 Chymotripsin-Like Protease (3CLPro) Activity, Foods, doi:10.3390/foods10071503.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
g.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
h.
Non-structural protein 10 (nsp10) serves as an RNA chaperone and stabilizes conformations of nsp12 and nsp14 in the replicase-transcriptase complex, which synthesizes new viral RNAs. Nsp10 disruption may destabilize replicase-transcriptase complex activity.
i.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
j.
The interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor is a primary method of viral entry, inhibiting this interaction can prevent the virus from attaching to and entering host cells, halting infection at an early stage.
k.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
l.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
m.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
n.
A549-AT is a human lung carcinoma cell line stably transfected with ACE2 and TMPRSS2 receptors. Unlike the parental line, this overexpression ensures stable infection and enhanced viral entry, allowing for the evaluation of antiviral efficacy against various SARS-CoV-2 variants.
o.
293T is a human embryonic kidney cell line that can be engineered for high ACE2 expression and SARS-CoV-2 susceptibility. 293T cells are easily transfected and support high protein expression.
p.
HEK293-hACE2 is a human embryonic kidney cell line with high ACE2 expression and SARS-CoV-2 susceptibility. Cells have been transfected with a plasmid to express the human ACE2 (hACE2) protein.
q.
293T/hACE2/TMPRSS2 is a human embryonic kidney cell line engineered for high ACE2 and TMPRSS2 expression, which mimics key aspects of human infection. 293T/hACE2/TMPRSS2 cells are very susceptible to SARS-CoV-2 infection.
r.
Vero E6 is an African green monkey kidney cell line with low/no ACE2 expression and high SARS-CoV-2 susceptibility. The cell line is easy to maintain and supports robust viral replication, however the monkey origin may not accurately represent human responses.
s.
SH-SY5Y is a human neuroblastoma cell line that exhibits neuronal phenotypes. It is commonly used as an in vitro model for studying neurotoxicity, neurodegenerative diseases, and neuronal differentiation.
Alici et al., 26 May 2025, peer-reviewed, 1 author.
Contact: hakanalici@beun.edu.tr.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Structure-Based Design and In-Silico Evaluation of Computationally Proposed Curcumin Derivatives as Potential Inhibitors of the Coronaviral PLpro Enzymes
Pharmaceuticals, doi:10.3390/ph18060798
Background/Objectives: Highly pathogenic coronaviruses (CoVs), including SARS-CoV, MERS-CoV, and SARS-CoV-2, continue to pose a significant threat to global public health. Therefore, this situation highlights the urgent need for effective broadspectrum antiviral agents. Curcumin, a naturally occurring polyphenol known for its antiviral and anti-inflammatory properties, faces limitations such as poor bioavailability and rapid metabolic degradation, restricting its practical therapeutic application. Methods: To address these limitations, this study introduces a novel design strategy aimed at 42 new curcumin derivatives with improved pharmacokinetic profiles, specifically targeting the conserved coronavirus enzyme papain-like protease (PLpro). A comprehensive in silico evaluation was performed, including ADMET (Absorption, Distribution, Metabolism, Elimination, and Toxicity) analysis, molecular docking, molecular dynamics (MD) simulations, and Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) calculations. Results: Extensive pharmacokinetic and toxicological assessments (ADMET analyses) identified 19 derivatives exhibiting optimal drug-like characteristics according to Lipinski's Rule of Five (Ro5). Molecular docking analyses demonstrated that these novel derivatives possess significantly enhanced binding affinities to PLpro enzymes from SARS-CoV, MERS-CoV, and SARS-CoV-2 compared to standard antiviral agents and natural curcumin. Further validation through MD simulations and MM/PBSA calculations confirmed the structural stability and robust interactions of the most promising derivatives within the SARS-CoV PLpro active site. Conclusions: The results of this study provide essential structural and functional insights, reinforcing the potential of these newly developed curcumin derivatives as potent, broad-spectrum antiviral agents effective against current and future coronavirus threats.
Supplementary Information (Figures S2-S7 ) for potential future experimental validation. Also, general structures of the final target compounds.
Institutional Review Board Statement: Not applicable. Informed Consent Statement: Not applicable.
Conflicts of Interest: The author declares no conflicts of interest.
References
Abraham, Murtola, Schulz, Páll, Smith et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, doi:10.1016/j.softx.2015.06.001
Ajavon, Bonate, Taft, Renal excretion of clofarabine: Assessment of dose-linearity and role of renal transport systems on drug excretion, Eur. J. Pharm. Sci, doi:10.1016/j.ejps.2010.03.014
Ali, Van Boheemen, Bestebroer Theo, Osterhaus Albert, Fouchier Ron, Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia, N. Engl. J. Med, doi:10.1056/NEJMoa1211721
Alici, Tahtaci, Demir, Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes, Comput. Biol. Chem, doi:10.1016/j.compbiolchem.2022.107657
Andersen, Rambaut, Lipkin, Holmes, Garry, The proximal origin of SARS-CoV-2, Nat. Med, doi:10.1038/s41591-020-0820-9
Andersen, Rattle: A "velocity" version of the shake algorithm for molecular dynamics calculations, J. Comput. Phys, doi:10.1016/0021-9991(83)90014-1
Armani, Geier, Forst, Merle, Alpers et al., Effect of changes in metabolic enzymes and transporters on drug metabolism in the context of liver disease: Impact on pharmacokinetics and drug-drug interactions, Br. J. Clin. Pharmacol, doi:10.1111/bcp.15990
Azarkar, Abedi, Lavasani, Ammameh, Goharipanah et al., Curcumin as a natural potential drug candidate against important zoonotic viruses and prions: A narrative review, Phytother. Res, doi:10.1002/ptr.8119
Bader, Calleja, Devine, Kuchel, Lu et al., A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID, Nat. Commun, doi:10.1038/s41467-025-57905-4
Baez-Santos, St John, Mesecar, The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds, Antivir. Res, doi:10.1016/j.antiviral.2014.12.015
Barretto, Jukneliene, Ratia, Chen, Mesecar et al., The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity, J. Virol, doi:10.1128/JVI.79.24.15189-15198.2005
Bastos, De Aguiar, Cruz, Ramos, Kimani et al., Rational Approach toward COVID-19 ′ s Main Protease Inhibitors: A Hierarchical Biochemoinformatics Analysis, Int. J. Mol. Sci, doi:10.3390/ijms25126715
Bormann, Alt, Schipper, Van De Sand, Le-Trilling et al., Turmeric Root and Its Bioactive Ingredient Curcumin Effectively Neutralize SARS-CoV-2 In Vitro, Viruses, doi:10.3390/v13101914
Bussi, Donadio, Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys, doi:10.1063/1.2408420
Calkilic, Alici, Direkel, Tahtaci, Synthesis, Characterization, Theoretical Analyses, and Investigation of Their Biological Activities of Acetovanillone-Derived Novel Benzyl Ethers, Polycycl. Aromat. Comp, doi:10.1080/10406638.2021.1950782
Chen, Lien, Chen, Hung, Lin et al., Synthesis and Evaluation of Novel Derivatives of Curcuminoids with Cytotoxicity, Int. J. Mol. Sci, doi:10.3390/ijms222212171
Cui, Li, Shi, Origin and evolution of pathogenic coronaviruses, Nat. Rev. Microbiol, doi:10.1038/s41579-018-0118-9
Da Silva Lopes, Pereira, Lima, Curcumin and Neurodegenerative Diseases: From Traditional to Translational Medicines
Dai, Zhang, Zheng, Luo, Chen et al., Advances in β-Diketocyclisation of Curcumin Derivatives and their Antitumor Activity, Chem. Biodivers, doi:10.1002/cbdv.202301556
Daina, Michielin, Zoete, SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Sci. Rep, doi:10.1038/srep42717
Darden, York, Pedersen, Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems, J. Chem. Phys, doi:10.1063/1.464397
Dei Cas, Ghidoni, Dietary Curcumin: Correlation between Bioavailability and Health Potential, Nutrients, doi:10.3390/nu11092147
Dourado, Freire, Pereira, Amaral-Machado, Alencar et al., Will curcumin nanosystems be the next promising antiviral alternatives in COVID-19 treatment trials?, Biomed. Pharmacother, doi:10.1016/j.biopha.2021.111578
Eltayb, Abdalla, Rabie, Novel Investigational Anti-SARS-CoV-2 Agent Ensitrelvir "S-217622": A Very Promising Potential Universal Broad-Spectrum Antiviral at the Therapeutic Frontline of Coronavirus Species, ACS Omega, doi:10.1021/acsomega.2c03881
Fakih, Ritmaleni; Zainul, Muchtaridi, Molecular docking-based virtual screening and computational investigations of biomolecules (curcumin analogs) as potential lead inhibitors for SARS-CoV-2 papain-like protease, Pharmacia, doi:10.3897/pharmacia.71.e123948
Farooqui, Metabolism, Bioavailability, Biochemical Effects of Curcumin in Visceral Organs and the Brain
Ferreira, Villanueva, Al Adem, Fadl, Alzyoud et al., Identification of novel allosteric sites of SARS-CoV-2 papain-like protease (PLpro) for the development of COVID-19 antivirals, J. Biol. Chem, doi:10.1016/j.jbc.2024.107821
Fibriani, Taharuddin, Stephanie, Yamahoki, Laurelia et al., Curcumin-derived carbon-dots as a potential COVID-19 antiviral drug, Heliyon, doi:10.1016/j.heliyon.2023.e20089
Garrido, Lepailleur, Mignani, Dallemagne, Rochais, hERG toxicity assessment: Useful guidelines for drug design, European J. Med. Chem, doi:10.1016/j.ejmech.2020.112290
Ghose, Viswanadhan, Wendoloski, A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases, J. Comb. Chem, doi:10.1021/cc9800071
Ghosh, Brindisi, Shahabi, Chapman, Mesecar, Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics, ChemMedChem, doi:10.1002/cmdc.202000223
Gold, Reis, Glaser, Glickman, Coronaviral PLpro proteases and the immunomodulatory roles of conjugated versus free Interferon Stimulated Gene product-15 (ISG15), Semin. Cell Dev. Biol, doi:10.1016/j.semcdb.2022.06.005
Gu, Zhang, Zhang, Wang, Sun et al., Unveiling the mechanism of action of a novel natural dual inhibitor of SARS-CoV-2 Mpro and PLpro with molecular dynamics simulations, Nat. Prod. Bioprospect, doi:10.1007/s13659-024-00486-4
Gupta, Prasad, Kim, Patchva, Webb et al., Multitargeting by curcumin as revealed by molecular interaction studies, Nat. Prod. Rep, doi:10.1039/c1np00051a
Guy, Saccharin, Encyclopedia of Toxicology
Hanwell, Curtis, Lonie, Vandermeersch, Zurek et al., An advanced semantic chemical editor, visualization, and analysis platform, J. Cheminform, doi:10.1186/1758-2946-4-17
Hilgers, Conradi, Burton, Caco-2 cell monolayers as a model for drug transport across the intestinal mucosa, Pharm. Res, doi:10.1023/A:1015937605100
Huang, Rauscher, Nawrocki, Ran, Feig et al., CHARMM36m: An improved force field for folded and intrinsically disordered proteins, Nat. Methods, doi:10.1038/nmeth.4067
Imane, Lamiae El, Oussama, Mohammed, Mhammed El et al., Integrated Exploration of Pyranocoumarin Derivatives as Synergistic Inhibitors of Dual-target for Mpro and PLpro Proteins of SARS-CoV-2 through Molecular Docking, ADMET Analysis, and Molecular Dynamics Simulation, Curr. Med. Chem
Jung, Goo, Hwang, Lee, Kim et al., Absorption Distribution Metabolism Excretion and Toxicity Property Prediction Utilizing a Pre-Trained Natural Language Processing Model and Its Applications in Early-Stage Drug Development, Pharmaceuticals, doi:10.3390/ph17030382
Knights, Stresser, Miners, Crespi, In Vitro Drug Metabolism Using Liver Microsomes, Curr. Protoc. Pharmacol, doi:10.1002/cpph.9
Kono, Kawahara, Shinozaki, Nomura, Marutani et al., Characterization of P-Glycoprotein Inhibitors for Evaluating the Effect of P-Glycoprotein on the Intestinal Absorption of Drugs, Pharmaceutics, doi:10.3390/pharmaceutics13030388
Korzekwa, Nagar, Drug Distribution Part 2. Predicting Volume of Distribution from Plasma Protein Binding and Membrane Partitioning, Pharm. Res, doi:10.1007/s11095-016-2086-y
Kumari, Kumar, Lynn, g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations, J. Chem. Inf. Model
Lee, Lei, Santarsiero, Gatuz, Cao et al., Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV, ACS Chem. Biol, doi:10.1021/cb500917m
Li, De Clercq, Therapeutic options for the 2019 novel coronavirus (2019-nCoV), Nat. Rev. Drug Discov, doi:10.1038/d41573-020-00016-0
Li, Edward, Guy, Drug-Like Property Concepts in Pharmaceutical Design, Curr. Pharm. Des
Lipinski, Lead-and drug-like compounds: The rule-of-five revolution, Drug Discov. Today Technol, doi:10.1016/j.ddtec.2004.11.007
Lipinski, Lombardo, Dominy, Feeney, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Deliv. Rev, doi:10.1016/S0169-409X(96)00423-1
Metwaly, Elkaeed, Khalifa, Alsfouk, Amin et al., Discovery of potential FDAapproved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach, J. Chem. Res, doi:10.1177/17475198241298547
Morris, Huey, Lindstrom, Sanner, Belew et al., AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem, doi:10.1002/jcc.21256
Mortelmans, Zeiger, The Ames Salmonella/microsome mutagenicity assay, Mutat. Res.-Fundam. Mol. Mech. Mutagen, doi:10.1016/S0027-5107(00)00064-6
Nandi, Kumar, Saxena, QSAR of SARS-CoV-2 Main Protease Inhibitors Utilizing Theoretical Molecular Descriptors, Lett. Drug Des. Discov, doi:10.2174/1570180820666221214151614
Nandi, Kumar, Saxena, Repurposing of Drugs and HTS to Combat SARS-CoV-2 Main Protease Utilizing Structure-Based Molecular Docking, Lett. Drug Des. Discov, doi:10.2174/1570180818666211007111105
Nicoliche, Bartolomeo, Lemes, Pereira, Nunes et al., anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2, Sci. Rep, doi:10.1038/s41598-024-61662-7
Nishizawa, Yoda, Morokado, Komori, Nakanishi et al., Changes of drug pharmacokinetics mediated by downregulation of kidney organic cation transporters Mate1 and Oct2 in a rat model of hyperuricemia, PLoS ONE, doi:10.1371/journal.pone.0214862
O'boyle, Banck, James, Morley, Vandermeersch et al., Open Babel: An open chemical toolbox, J. Cheminform, doi:10.1186/1758-2946-3-33
O'hagan, Kell, The apparent permeabilities of Caco-2 cells to marketed drugs: Magnitude, and independence from both biophysical properties and endogenite similarities, PeerJ, doi:10.7717/peerj.1405
Ozcan, Akkoc, Alici, Capanlar, Sahin et al., Novel Thioether-Bridged 2,6-Disubstituted and 2,5,6-Trisubstituted Imidazothiadiazole Analogues: Synthesis, Antiproliferative Activity, ADME, and Molecular Docking Studies, Chem. Biodivers, doi:10.1002/cbdv.202200884
Ozcan, Alici, Taslimi, Tahtaci, Novel 1,2,4-triazole-derived Schiff base derivatives: Design, synthesis, and multienzyme targeting potential for therapeutic applications, Bioorg. Chem, doi:10.1016/j.bioorg.2025.108246
Parrinello, Rahman, Polymorphic transitions in single crystals: A new molecular dynamics method, J. Appl. Phys, doi:10.1063/1.328693
Peiris, Chu, Cheng, Chan, Hung et al., Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study, Lancet, doi:10.1016/S0140-6736(03)13412-5
Pelkonen, Boobis, Gundert-Remy, In vitro prediction of gastrointestinal absorption and bioavailability: An experts' meeting report, Eur. J. Clin. Pharmacol, doi:10.1007/s002280100369
Pillai, Dhanikula, Panchagnula, Drug delivery: An odyssey of 100 years, Curr. Opin. Chem. Biol, doi:10.1016/S1367-5931(00)00226-X
Pires, Blundell, Ascher, Pkcsm, Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures, J. Med. Chem, doi:10.1021/acs.jmedchem.5b00104
Prasad, Tyagi, Aggarwal, Recent Developments in Delivery, Bioavailability, Absorption and Metabolism of Curcumin: The Golden Pigment from Golden Spice, Cancer Res. Treat, doi:10.4143/crt.2014.46.1.2
Rabie, Abdalla, Evaluation of a series of nucleoside analogs as effective anticoronaviral-2 drugs against the Omicron-B.1.1.529/BA.2 subvariant: A repurposing research study, Med. Chem. Res, doi:10.1007/s00044-022-02970-3
Rabie, Abdalla, Forodesine and Riboprine Exhibit Strong Anti-SARS-CoV-2 Repurposing Potential: In Silico and In Vitro Studies, ACS Bio Med. Chem. Au, doi:10.1021/acsbiomedchemau.2c00039
Rabie, Efficacious Preclinical Repurposing of the Nucleoside Analogue Didanosine against COVID-19 Polymerase and Exonuclease, ACS Omega, doi:10.1021/acsomega.1c07095
Rabie, Eltayb, Potent Dual Polymerase/Exonuclease Inhibitory Activities of Antioxidant Aminothiadiazoles Against the COVID-19 Omicron Virus: A Promising In Silico/In Vitro Repositioning Research Study, Mol. Biotechnol, doi:10.1007/s12033-022-00551-8
Rabie, Future of the current anticoronaviral agents: A viewpoint on the validation for the next COVIDs and pandemics, Biocell, doi:10.32604/biocell.2023.030057
Rabie, Improved synthesis of the anti-SARS-CoV-2 investigational agent (E)-N-(4-cyanobenzylidene)-6-fluoro-3hydroxypyrazine-2-carboxamide (cyanorona-20), Rev. Chim, doi:10.37358/RC.22.4.8555
Rabie, Khedraoui, Chtita, Targeting Conserved Regions of the SARS-CoV-2 Polymerase (RdRp) with Kinase Inhibitors as an Effective New Tactic for Discovering Dual-Action "Antiviral-Antiinflammatory" Drugs against COVID-19, Comput. Biol. Chem, doi:10.1016/j.compbiolchem.2025.108454
Rabie, New Potential Inhibitors of Coronaviral Main Protease (CoV-Mpro): Strychnine Bush, Pineapple, and Ginger could be Natural Enemies of COVID-19, Int. J. New Chem
Rabie, Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication, ACS Omega, doi:10.1021/acsomega.1c05998
Rabie, Potent toxic effects of Taroxaz-104 on the replication of SARS-CoV-2 particles, Chem.-Biol. Interact, doi:10.1016/j.cbi.2021.109480
Rabie, RNA: The most attractive target in recent viral diseases, Chem. Biol. Drug Des, doi:10.1111/cbdd.14404
Rabie, Revolutionizing Playing with Skeleton Atoms: Molecular Editing Surgery in Medicinal Chemistry, Mini Rev. Med. Chem, doi:10.2174/0113895575316229240611113946
Rabie, Teriflunomide: A possible effective drug for the comprehensive treatment of COVID-19, Curr. Res. Pharmacol. Drug Discov, doi:10.1016/j.crphar.2021.100055
Rabie, The informative nature of the disappeared SARS-CoV-2 genomic sequences: A mini-review with perspectives, Adv. Chemicobiol. Res, doi:10.37256/acbr.1220221403
Rabie, Two antioxidant 2,5-disubstituted-1,3,4-oxadiazoles (CoViTris2020 and ChloViD2020): Successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs, New J. Chem, doi:10.1039/D0NJ03708G
Rabie, Yamari, Chtita, The isoquinoline derivative "CYNOVID" as a prospective anti-SARS-CoV-2 agent: An expanded investigative computational study, Eur. J. Med. Chem. Rep, doi:10.1016/j.ejmcr.2024.100214
Rai, Feitosa, Springer Nature Singapore
Ratia, Saikatendu, Santarsiero, Barretto, Baker et al., Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0510851103
Rut, Lv, Zmudzinski, Patchett, Nayak et al., Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design, Sci. Adv, doi:10.1126/sciadv.abd4596
Severance, Sandoval, Wright, Correlation between Apparent Substrate Affinity and OCT2 Transport Turnover, J. Pharmacol. Exp. Ther, doi:10.1124/jpet.117.242552
Shin, Mukherjee, Grewe, Bojkova, Baek et al., Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity, Nature, doi:10.1038/s41586-020-2601-5
Shyr, Gorshkov, Chen, Zheng, Drug Discovery Strategies for SARS-CoV-2, J. Pharmacol. Exp. Ther, doi:10.1124/jpet.120.000123
Silvestre, Santos, Silva, Ombredane, Pinheiro et al., Pharmacokinetics of Curcumin Delivered by Nanoparticles and the Relationship with Antitumor Efficacy: A Systematic Review, Pharmaceuticals, doi:10.3390/ph16070943
Siviero, Gallo, Maggini, Gori, Mugelli et al., Curcumin, a golden spice with a low bioavailability, J. Herb. Med, doi:10.1016/j.hermed.2015.03.001
Smith, Jones, Walker, Design of Drugs Involving the Concepts and Theories of Drug Metabolism and Pharmacokinetics, Med. Res. Rev, doi:10.1002/(SICI)1098-1128(199605)16:3%3C243::AID-MED2%3E3.0.CO;2-Z
Stepensky, Use of unbound volumes of drug distribution in pharmacokinetic calculations, Eur. J. Pharm. Sci, doi:10.1016/j.ejps.2010.10.011
Stillhart, Vučićević, Augustijns, Basit, Batchelor et al., Impact of gastrointestinal physiology on drug absorption in special populations-An UNGAP review, Eur. J. Pharm. Sci, doi:10.1016/j.ejps.2020.105280
Suresh, Nangia, Curcumin: Pharmaceutical solids as a platform to improve solubility and bioavailability, CrystEngComm, doi:10.1039/C8CE00469B
Suzuki, Taniyama, Aoyama, Watanabe, Evaluation of the Role of P-glycoprotein (P-gp)-Mediated Efflux in the Intestinal Absorption of Common Substrates with Elacridar, a P-gp Inhibitor, in Rats, Eur. J. Drug Metab. Pharmacokinet, doi:10.1007/s13318-019-00602-7
Szakács, Váradi, Özvegy-Laczka, Sarkadi, The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox), Drug Discov. Today, doi:10.1016/j.drudis.2007.12.010
Teixeira, Medeiros, Da Silva Oliveira, Acha, Pereira-Freire, Effect of Curcumin on the Process of Neuroinflammation Caused by COVID-19
Teshima, Takeshi, Zhengmao, Ken, Takao et al., Antiviral activity of curcumin and its analogs selected by an artificial intelligence-supported activity prediction system in SARS-CoV-2-infected VeroE6 cells, Nat. Prod. Res, doi:10.1080/14786419.2023.2194647
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem, doi:10.1002/jcc.21334
Valdés-Albuernes, Díaz-Pico, Alfaro, Caballero, Modeling of noncovalent inhibitors of the papain-like protease (PLpro) from SARS-CoV-2 considering the protein flexibility by using molecular dynamics and cross-docking, Front. Mol. Biosci, doi:10.3389/fmolb.2024.1374364
Van Vliet, Huynh, Palà, Patel, Singer et al., Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site, PLoS Pathog, doi:10.1371/journal.ppat.1011065
Vanommeslaeghe, Hatcher, Acharya, Kundu, Zhong et al., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comput. Chem, doi:10.1002/jcc.21367
Vanommeslaeghe, Mackerell, Jr, Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing, J. Chem. Inf. Model, doi:10.1021/ci300363c
Varghese, Liu, Liu, Guo, Dong et al., Analysis of Structures of SARS-CoV-2 Papain-like Protease Bound with Ligands Unveils Structural Features for Inhibiting the Enzyme, Molecules, doi:10.3390/molecules30030491
Veber, Johnson, Cheng, Smith, Ward et al., Molecular Properties That Influence the Oral Bioavailability of Drug Candidates, J. Med. Chem, doi:10.1021/jm020017n
Waring, Lipophilicity in drug discovery, Expert. Opin. Drug Discov, doi:10.1517/17460441003605098
Waters, Lombardo, Use of the Øie-Tozer Model in Understanding Mechanisms and Determinants of Drug Distribution, Drug Metab. Dispos, doi:10.1124/dmd.110.032458
Wei, Senanayake, Bohling, Vinogradov, Targeted Nanogel Conjugate for Improved Stability and Cellular Permeability of Curcumin: Synthesis, Pharmacokinetics, and Tumor Growth Inhibition, Mol. Pharm, doi:10.1021/mp500290f
Wu, Zhao, Yu, Chen, Wang et al., A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Xiong, Wu, Yi, Fu, Yang et al., 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties, Nucleic Acids Res, doi:10.1002/minf.201500040
Yang, Lou, Sun, Li, Cai et al., admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties, Bioinformatics, doi:10.1093/bioinformatics/bty707
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zumla, Chan, Azhar, Hui, Yuen, Coronaviruses-Drug discovery and therapeutic options, Nat. Rev. Drug Discov, doi:10.1038/nrd.2015.37
Çevik, Işik, Karakaya, ADMET and Physicochemical Assessments in Drug Design
DOI record:
{
"DOI": "10.3390/ph18060798",
"ISSN": [
"1424-8247"
],
"URL": "http://dx.doi.org/10.3390/ph18060798",
"abstract": "<jats:p>Background/Objectives: Highly pathogenic coronaviruses (CoVs), including SARS-CoV, MERS-CoV, and SARS-CoV-2, continue to pose a significant threat to global public health. Therefore, this situation highlights the urgent need for effective broad-spectrum antiviral agents. Curcumin, a naturally occurring polyphenol known for its antiviral and anti-inflammatory properties, faces limitations such as poor bioavailability and rapid metabolic degradation, restricting its practical therapeutic application. Methods: To address these limitations, this study introduces a novel design strategy aimed at 42 new curcumin derivatives with improved pharmacokinetic profiles, specifically targeting the conserved coronavirus enzyme papain-like protease (PLpro). A comprehensive in silico evaluation was performed, including ADMET (Absorption, Distribution, Metabolism, Elimination, and Toxicity) analysis, molecular docking, molecular dynamics (MD) simulations, and Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) calculations. Results: Extensive pharmacokinetic and toxicological assessments (ADMET analyses) identified 19 derivatives exhibiting optimal drug-like characteristics according to Lipinski’s Rule of Five (Ro5). Molecular docking analyses demonstrated that these novel derivatives possess significantly enhanced binding affinities to PLpro enzymes from SARS-CoV, MERS-CoV, and SARS-CoV-2 compared to standard antiviral agents and natural curcumin. Further validation through MD simulations and MM/PBSA calculations confirmed the structural stability and robust interactions of the most promising derivatives within the SARS-CoV PLpro active site. Conclusions: The results of this study provide essential structural and functional insights, reinforcing the potential of these newly developed curcumin derivatives as potent, broad-spectrum antiviral agents effective against current and future coronavirus threats.</jats:p>",
"alternative-id": [
"ph18060798"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0001-5105-8331",
"affiliation": [
{
"name": "Department of Physics, Faculty of Science, Zonguldak Bülent Ecevit University, 67100 Zonguldak, Türkiye"
}
],
"authenticated-orcid": false,
"family": "Alici",
"given": "Hakan",
"sequence": "first"
}
],
"container-title": "Pharmaceuticals",
"container-title-short": "Pharmaceuticals",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
5,
29
]
],
"date-time": "2025-05-29T08:46:38Z",
"timestamp": 1748508398000
},
"deposited": {
"date-parts": [
[
2025,
5,
29
]
],
"date-time": "2025-05-29T08:56:04Z",
"timestamp": 1748508964000
},
"indexed": {
"date-parts": [
[
2025,
5,
30
]
],
"date-time": "2025-05-30T04:05:02Z",
"timestamp": 1748577902436,
"version": "3.41.0"
},
"is-referenced-by-count": 0,
"issue": "6",
"issued": {
"date-parts": [
[
2025,
5,
26
]
]
},
"journal-issue": {
"issue": "6",
"published-online": {
"date-parts": [
[
2025,
6
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
5,
26
]
],
"date-time": "2025-05-26T00:00:00Z",
"timestamp": 1748217600000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1424-8247/18/6/798/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "798",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
5,
26
]
]
},
"published-online": {
"date-parts": [
[
2025,
5,
26
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/s41579-018-0118-9",
"article-title": "Origin and evolution of pathogenic coronaviruses",
"author": "Cui",
"doi-asserted-by": "crossref",
"first-page": "181",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_1",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A pneumonia outbreak associated with a new coronavirus of probable bat origin",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nature",
"key": "ref_2",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(03)13412-5",
"article-title": "Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study",
"author": "Peiris",
"doi-asserted-by": "crossref",
"first-page": "1767",
"journal-title": "Lancet",
"key": "ref_3",
"volume": "361",
"year": "2003"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"article-title": "A new coronavirus associated with human respiratory disease in China",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "265",
"journal-title": "Nature",
"key": "ref_4",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa1211721",
"article-title": "Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia",
"doi-asserted-by": "crossref",
"first-page": "1814",
"journal-title": "N. Engl. J. Med.",
"key": "ref_5",
"volume": "367",
"year": "2012"
},
{
"DOI": "10.1038/nrd.2015.37",
"article-title": "Coronaviruses—Drug discovery and therapeutic options",
"author": "Zumla",
"doi-asserted-by": "crossref",
"first-page": "327",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "ref_6",
"volume": "15",
"year": "2016"
},
{
"DOI": "10.1038/d41573-020-00016-0",
"article-title": "Therapeutic options for the 2019 novel coronavirus (2019-nCoV)",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "149",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "ref_7",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1124/jpet.120.000123",
"article-title": "Drug Discovery Strategies for SARS-CoV-2",
"author": "Shyr",
"doi-asserted-by": "crossref",
"first-page": "127",
"journal-title": "J. Pharmacol. Exp. Ther.",
"key": "ref_8",
"volume": "375",
"year": "2020"
},
{
"article-title": "The informative nature of the disappeared SARS-CoV-2 genomic sequences: A mini-review with perspectives",
"author": "Rabie",
"first-page": "58",
"journal-title": "Adv. Chemicobiol. Res.",
"key": "ref_9",
"volume": "1",
"year": "2022"
},
{
"DOI": "10.1002/cmdc.202000223",
"article-title": "Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics",
"author": "Ghosh",
"doi-asserted-by": "crossref",
"first-page": "907",
"journal-title": "ChemMedChem",
"key": "ref_10",
"volume": "15",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-0820-9",
"article-title": "The proximal origin of SARS-CoV-2",
"author": "Andersen",
"doi-asserted-by": "crossref",
"first-page": "450",
"journal-title": "Nat. Med.",
"key": "ref_11",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2014.12.015",
"article-title": "The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds",
"author": "Mesecar",
"doi-asserted-by": "crossref",
"first-page": "21",
"journal-title": "Antivir. Res.",
"key": "ref_12",
"volume": "115",
"year": "2015"
},
{
"DOI": "10.1126/sciadv.abd4596",
"article-title": "Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design",
"author": "Rut",
"doi-asserted-by": "crossref",
"first-page": "eabd4596",
"journal-title": "Sci. Adv.",
"key": "ref_13",
"volume": "6",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2601-5",
"article-title": "Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity",
"author": "Shin",
"doi-asserted-by": "crossref",
"first-page": "657",
"journal-title": "Nature",
"key": "ref_14",
"volume": "587",
"year": "2020"
},
{
"DOI": "10.1128/JVI.79.24.15189-15198.2005",
"article-title": "The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity",
"author": "Barretto",
"doi-asserted-by": "crossref",
"first-page": "15189",
"journal-title": "J. Virol.",
"key": "ref_15",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1073/pnas.0510851103",
"article-title": "Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme",
"author": "Ratia",
"doi-asserted-by": "crossref",
"first-page": "5717",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_16",
"volume": "103",
"year": "2006"
},
{
"DOI": "10.1371/journal.ppat.1011065",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "van Vliet, V.J.E., Huynh, N., Palà, J., Patel, A., Singer, A., Slater, C., Chung, J., van Huizen, M., Teyra, J., and Miersch, S. (2022). Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site. PLoS Pathog., 18."
},
{
"DOI": "10.1016/j.semcdb.2022.06.005",
"article-title": "Coronaviral PLpro proteases and the immunomodulatory roles of conjugated versus free Interferon Stimulated Gene product-15 (ISG15)",
"author": "Gold",
"doi-asserted-by": "crossref",
"first-page": "16",
"journal-title": "Semin. Cell Dev. Biol.",
"key": "ref_18",
"volume": "132",
"year": "2022"
},
{
"DOI": "10.3390/molecules30030491",
"doi-asserted-by": "crossref",
"key": "ref_19",
"unstructured": "Varghese, A., Liu, J., Liu, B., Guo, W., Dong, F., Patterson, T.A., and Hong, H. (2025). Analysis of Structures of SARS-CoV-2 Papain-like Protease Bound with Ligands Unveils Structural Features for Inhibiting the Enzyme. Molecules, 30."
},
{
"DOI": "10.1016/j.compbiolchem.2022.107657",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Alici, H., Tahtaci, H., and Demir, K. (2022). Design and various in silico studies of the novel curcumin derivatives as potential candidates against COVID-19 -associated main enzymes. Comput. Biol. Chem., 98."
},
{
"DOI": "10.1007/978-981-99-7731-4",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Rai, M., and Feitosa, C.M. (2023). Effect of Curcumin on the Process of Neuroinflammation Caused by COVID-19. Curcumin and Neurodegenerative Diseases: From Traditional to Translational Medicines, Springer Nature Singapore."
},
{
"DOI": "10.1007/978-3-319-15889-1_3",
"doi-asserted-by": "crossref",
"key": "ref_22",
"unstructured": "Farooqui, A.A. (2016). Metabolism, Bioavailability, Biochemical Effects of Curcumin in Visceral Organs and the Brain. Therapeutic Potentials of Curcumin for Alzheimer Disease, Springer International Publishing."
},
{
"DOI": "10.4143/crt.2014.46.1.2",
"article-title": "Recent Developments in Delivery, Bioavailability, Absorption and Metabolism of Curcumin: The Golden Pigment from Golden Spice",
"author": "Prasad",
"doi-asserted-by": "crossref",
"first-page": "2",
"journal-title": "Cancer Res. Treat.",
"key": "ref_23",
"volume": "46",
"year": "2014"
},
{
"DOI": "10.1016/j.hermed.2015.03.001",
"article-title": "Curcumin, a golden spice with a low bioavailability",
"author": "Siviero",
"doi-asserted-by": "crossref",
"first-page": "57",
"journal-title": "J. Herb. Med.",
"key": "ref_24",
"volume": "5",
"year": "2015"
},
{
"DOI": "10.1039/C8CE00469B",
"article-title": "Curcumin: Pharmaceutical solids as a platform to improve solubility and bioavailability",
"author": "Suresh",
"doi-asserted-by": "crossref",
"first-page": "3277",
"journal-title": "CrystEngComm",
"key": "ref_25",
"volume": "20",
"year": "2018"
},
{
"DOI": "10.1080/14786419.2023.2194647",
"article-title": "Antiviral activity of curcumin and its analogs selected by an artificial intelligence-supported activity prediction system in SARS-CoV-2-infected VeroE6 cells",
"author": "Teshima",
"doi-asserted-by": "crossref",
"first-page": "867",
"journal-title": "Nat. Prod. Res.",
"key": "ref_26",
"volume": "38",
"year": "2024"
},
{
"DOI": "10.1016/j.heliyon.2023.e20089",
"article-title": "Curcumin-derived carbon-dots as a potential COVID-19 antiviral drug",
"author": "Fibriani",
"doi-asserted-by": "crossref",
"first-page": "e20089",
"journal-title": "Heliyon",
"key": "ref_27",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1016/j.biopha.2021.111578",
"doi-asserted-by": "crossref",
"key": "ref_28",
"unstructured": "Dourado, D., Freire, D.T., Pereira, D.T., Amaral-Machado, L., Alencar, É.N., de Barros, A.L.B., and Egito, E.S.T. (2021). Will curcumin nanosystems be the next promising antiviral alternatives in COVID-19 treatment trials?. Biomed. Pharmacother., 139."
},
{
"DOI": "10.3390/v13101914",
"doi-asserted-by": "crossref",
"key": "ref_29",
"unstructured": "Bormann, M., Alt, M., Schipper, L., van de Sand, L., Le-Trilling, V.T., Rink, L., Heinen, N., Madel, R.J., Otte, M., and Wuensch, K. (2021). Turmeric Root and Its Bioactive Ingredient Curcumin Effectively Neutralize SARS-CoV-2 In Vitro. Viruses, 13."
},
{
"DOI": "10.1038/s41598-024-61662-7",
"doi-asserted-by": "crossref",
"key": "ref_30",
"unstructured": "Nicoliche, T., Bartolomeo, C.S., Lemes, R.M.R., Pereira, G.C., Nunes, T.A., Oliveira, R.B., Nicastro, A.L.M., Soares, É.N., da Cunha Lima, B.F., and Rodrigues, B.M. (2024). Antiviral, anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2. Sci. Rep., 14."
},
{
"DOI": "10.1002/ptr.8119",
"article-title": "Curcumin as a natural potential drug candidate against important zoonotic viruses and prions: A narrative review",
"author": "Azarkar",
"doi-asserted-by": "crossref",
"first-page": "3080",
"journal-title": "Phytother. Res.",
"key": "ref_31",
"volume": "38",
"year": "2024"
},
{
"article-title": "Molecular docking-based virtual screening and computational investigations of biomolecules (curcumin analogs) as potential lead inhibitors for SARS-CoV-2 papain-like protease",
"author": "Fakih",
"first-page": "1",
"journal-title": "Pharmacia",
"key": "ref_32",
"volume": "71",
"year": "2024"
},
{
"DOI": "10.1101/2023.05.16.540953",
"doi-asserted-by": "crossref",
"key": "ref_33",
"unstructured": "Ferreira, J.C., Villanueva, A.J., Al Adem, K., Fadl, S., Alzyoud, L., Ghattas, M.A., and Rabeh, W.M. (2024). Identification of novel allosteric sites of SARS-CoV-2 papain-like protease (PLpro) for the development of COVID-19 antivirals. J. Biol. Chem., 300."
},
{
"DOI": "10.1111/cbdd.14404",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Rabie, A.M. (2024). RNA: The most attractive target in recent viral diseases. Chem. Biol. Drug Des., 103."
},
{
"DOI": "10.1016/j.compbiolchem.2025.108454",
"doi-asserted-by": "crossref",
"key": "ref_35",
"unstructured": "Rabie, A.M., Khedraoui, M., and Chtita, S. (2025). Targeting Conserved Regions of the SARS-CoV-2 Polymerase (RdRp) with Kinase Inhibitors as an Effective New Tactic for Discovering Dual-Action “Antiviral─Antiinflammatory” Drugs against COVID-19. Comput. Biol. Chem., in press."
},
{
"DOI": "10.32604/biocell.2023.030057",
"article-title": "Future of the current anticoronaviral agents: A viewpoint on the validation for the next COVIDs and pandemics",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "2133",
"journal-title": "Biocell",
"key": "ref_36",
"volume": "47",
"year": "2023"
},
{
"article-title": "The isoquinoline derivative “CYNOVID” as a prospective anti-SARS-CoV-2 agent: An expanded investigative computational study",
"author": "Rabie",
"first-page": "100214",
"journal-title": "Eur. J. Med. Chem. Rep.",
"key": "ref_37",
"volume": "12",
"year": "2024"
},
{
"DOI": "10.1021/acsomega.1c05998",
"article-title": "Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "2960",
"journal-title": "ACS Omega",
"key": "ref_38",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1021/acsbiomedchemau.2c00039",
"article-title": "Forodesine and Riboprine Exhibit Strong Anti-SARS-CoV-2 Repurposing Potential: In Silico and In Vitro Studies",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "565",
"journal-title": "ACS Bio Med. Chem. Au",
"key": "ref_39",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.1016/j.crphar.2021.100055",
"article-title": "Teriflunomide: A possible effective drug for the comprehensive treatment of COVID-19",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "100055",
"journal-title": "Curr. Res. Pharmacol. Drug Discov.",
"key": "ref_40",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.1021/acsomega.2c03881",
"article-title": "Novel Investigational Anti-SARS-CoV-2 Agent Ensitrelvir “S-217622”: A Very Promising Potential Universal Broad-Spectrum Antiviral at the Therapeutic Frontline of Coronavirus Species",
"author": "Eltayb",
"doi-asserted-by": "crossref",
"first-page": "5234",
"journal-title": "ACS Omega",
"key": "ref_41",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1007/s00044-022-02970-3",
"article-title": "Evaluation of a series of nucleoside analogs as effective anticoronaviral-2 drugs against the Omicron-B.1.1.529/BA.2 subvariant: A repurposing research study",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "326",
"journal-title": "Med. Chem. Res.",
"key": "ref_42",
"volume": "32",
"year": "2023"
},
{
"DOI": "10.1007/s12033-022-00551-8",
"article-title": "Potent Dual Polymerase/Exonuclease Inhibitory Activities of Antioxidant Aminothiadiazoles Against the COVID-19 Omicron Virus: A Promising In Silico/In Vitro Repositioning Research Study",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "592",
"journal-title": "Mol. Biotechnol.",
"key": "ref_43",
"volume": "66",
"year": "2024"
},
{
"DOI": "10.1021/acsomega.1c07095",
"article-title": "Efficacious Preclinical Repurposing of the Nucleoside Analogue Didanosine against COVID-19 Polymerase and Exonuclease",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "21385",
"journal-title": "ACS Omega",
"key": "ref_44",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1016/j.cbi.2021.109480",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Rabie, A.M. (2021). Potent toxic effects of Taroxaz-104 on the replication of SARS-CoV-2 particles. Chem.-Biol. Interact., 343."
},
{
"DOI": "10.2174/1570180818666211007111105",
"article-title": "Repurposing of Drugs and HTS to Combat SARS-CoV-2 Main Protease Utilizing Structure-Based Molecular Docking",
"author": "Nandi",
"doi-asserted-by": "crossref",
"first-page": "413",
"journal-title": "Lett. Drug Des. Discov.",
"key": "ref_46",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.2174/1570180820666221214151614",
"article-title": "QSAR of SARS-CoV-2 Main Protease Inhibitors Utilizing Theoretical Molecular Descriptors",
"author": "Nandi",
"doi-asserted-by": "crossref",
"first-page": "116",
"journal-title": "Lett. Drug Des. Discov.",
"key": "ref_47",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.37358/RC.22.4.8555",
"article-title": "Improved synthesis of the anti-SARS-CoV-2 investigational agent (E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide (cyanorona-20)",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "69",
"journal-title": "Rev. Chim.",
"key": "ref_48",
"volume": "73",
"year": "2022"
},
{
"DOI": "10.1039/D0NJ03708G",
"article-title": "Two antioxidant 2,5-disubstituted-1,3,4-oxadiazoles (CoViTris2020 and ChloViD2020): Successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "761",
"journal-title": "New J. Chem.",
"key": "ref_49",
"volume": "45",
"year": "2021"
},
{
"article-title": "New Potential Inhibitors of Coronaviral Main Protease (CoV-Mpro): Strychnine Bush, Pineapple, and Ginger could be Natural Enemies of COVID-19",
"author": "Rabie",
"first-page": "225",
"journal-title": "Int. J. New Chem.",
"key": "ref_50",
"volume": "9",
"year": "2022"
},
{
"DOI": "10.1002/(SICI)1098-1128(199605)16:3<243::AID-MED2>3.0.CO;2-Z",
"article-title": "Design of Drugs Involving the Concepts and Theories of Drug Metabolism and Pharmacokinetics",
"author": "Smith",
"doi-asserted-by": "crossref",
"first-page": "243",
"journal-title": "Med. Res. Rev.",
"key": "ref_51",
"volume": "16",
"year": "1996"
},
{
"DOI": "10.1002/9781394249190.ch6",
"doi-asserted-by": "crossref",
"key": "ref_52",
"unstructured": "Çevik, U.A., Işik, A., and Karakaya, A. (2025). ADMET and Physicochemical Assessments in Drug Design. Computational Methods for Rational Drug Design, Wiley."
},
{
"DOI": "10.1016/S1367-5931(00)00226-X",
"article-title": "Drug delivery: An odyssey of 100 years",
"author": "Pillai",
"doi-asserted-by": "crossref",
"first-page": "439",
"journal-title": "Curr. Opin. Chem. Biol.",
"key": "ref_53",
"volume": "5",
"year": "2001"
},
{
"DOI": "10.1080/10406638.2021.1950782",
"article-title": "Synthesis, Characterization, Theoretical Analyses, and Investigation of Their Biological Activities of Acetovanillone-Derived Novel Benzyl Ethers",
"author": "Calkilic",
"doi-asserted-by": "crossref",
"first-page": "5671",
"journal-title": "Polycycl. Aromat. Comp.",
"key": "ref_54",
"volume": "42",
"year": "2022"
},
{
"DOI": "10.1002/cbdv.202200884",
"doi-asserted-by": "crossref",
"key": "ref_55",
"unstructured": "Ozcan, I., Akkoc, S., Alici, H., Capanlar, S., Sahin, O., and Tahtaci, H. (2023). Novel Thioether-Bridged 2,6-Disubstituted and 2,5,6-Trisubstituted Imidazothiadiazole Analogues: Synthesis, Antiproliferative Activity, ADME, and Molecular Docking Studies. Chem. Biodivers., 20."
},
{
"DOI": "10.1016/j.bioorg.2025.108246",
"doi-asserted-by": "crossref",
"key": "ref_56",
"unstructured": "Ozcan, I., Alici, H., Taslimi, P., and Tahtaci, H. (2025). Novel 1,2,4-triazole-derived Schiff base derivatives: Design, synthesis, and multi-enzyme targeting potential for therapeutic applications. Bioorg. Chem., 157."
},
{
"DOI": "10.3390/ph17030382",
"doi-asserted-by": "crossref",
"key": "ref_57",
"unstructured": "Jung, W., Goo, S., Hwang, T., Lee, H., Kim, Y.-K., Chae, J.-w., Yun, H.-y., and Jung, S. (2024). Absorption Distribution Metabolism Excretion and Toxicity Property Prediction Utilizing a Pre-Trained Natural Language Processing Model and Its Applications in Early-Stage Drug Development. Pharmaceuticals, 17."
},
{
"DOI": "10.1016/S0169-409X(96)00423-1",
"article-title": "Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings",
"author": "Lipinski",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Adv. Drug Deliv. Rev.",
"key": "ref_58",
"volume": "23",
"year": "1997"
},
{
"DOI": "10.1016/j.ddtec.2004.11.007",
"article-title": "Lead- and drug-like compounds: The rule-of-five revolution",
"author": "Lipinski",
"doi-asserted-by": "crossref",
"first-page": "337",
"journal-title": "Drug Discov. Today Technol.",
"key": "ref_59",
"volume": "1",
"year": "2004"
},
{
"DOI": "10.1007/978-981-99-7731-4",
"doi-asserted-by": "crossref",
"key": "ref_60",
"unstructured": "Rai, M., and Feitosa, C.M. (2023). Pharmacokinetics and Pharmacodynamics of Curcumin. Curcumin and Neurodegenerative Diseases: From Traditional to Translational Medicines, Springer Nature Singapore."
},
{
"DOI": "10.3390/nu11092147",
"doi-asserted-by": "crossref",
"key": "ref_61",
"unstructured": "Dei Cas, M., and Ghidoni, R. (2019). Dietary Curcumin: Correlation between Bioavailability and Health Potential. Nutrients, 11."
},
{
"DOI": "10.1021/mp500290f",
"article-title": "Targeted Nanogel Conjugate for Improved Stability and Cellular Permeability of Curcumin: Synthesis, Pharmacokinetics, and Tumor Growth Inhibition",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "3112",
"journal-title": "Mol. Pharm.",
"key": "ref_62",
"volume": "11",
"year": "2014"
},
{
"DOI": "10.3390/ph16070943",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Silvestre, F., Santos, C., Silva, V., Ombredane, A., Pinheiro, W., Andrade, L., Garcia, M., Pacheco, T., Joanitti, G., and Luz, G. (2023). Pharmacokinetics of Curcumin Delivered by Nanoparticles and the Relationship with Antitumor Efficacy: A Systematic Review. Pharmaceuticals, 16."
},
{
"DOI": "10.3390/ijms222212171",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Chen, C.-Y., Lien, J.-C., Chen, C.-Y., Hung, C.-C., and Lin, H.-C. (2021). Design, Synthesis and Evaluation of Novel Derivatives of Curcuminoids with Cytotoxicity. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1039/c1np00051a",
"article-title": "Multitargeting by curcumin as revealed by molecular interaction studies",
"author": "Gupta",
"doi-asserted-by": "crossref",
"first-page": "1937",
"journal-title": "Nat. Prod. Rep.",
"key": "ref_65",
"volume": "28",
"year": "2011"
},
{
"DOI": "10.1002/cbdv.202301556",
"doi-asserted-by": "crossref",
"key": "ref_66",
"unstructured": "Dai, H., Zhang, S., Zheng, X., Luo, Z., Chen, H., and Yao, X. (2024). Advances in β-Diketocyclisation of Curcumin Derivatives and their Antitumor Activity. Chem. Biodivers., 21."
},
{
"DOI": "10.2174/0113895575316229240611113946",
"article-title": "Revolutionizing Playing with Skeleton Atoms: Molecular Editing Surgery in Medicinal Chemistry",
"author": "Rabie",
"doi-asserted-by": "crossref",
"first-page": "190",
"journal-title": "Mini Rev. Med. Chem.",
"key": "ref_67",
"volume": "25",
"year": "2025"
},
{
"DOI": "10.1021/cc9800071",
"article-title": "A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases",
"author": "Ghose",
"doi-asserted-by": "crossref",
"first-page": "55",
"journal-title": "J. Comb. Chem.",
"key": "ref_68",
"volume": "1",
"year": "1999"
},
{
"DOI": "10.1021/jm020017n",
"article-title": "Molecular Properties That Influence the Oral Bioavailability of Drug Candidates",
"author": "Veber",
"doi-asserted-by": "crossref",
"first-page": "2615",
"journal-title": "J. Med. Chem.",
"key": "ref_69",
"volume": "45",
"year": "2002"
},
{
"DOI": "10.1016/j.drudis.2007.12.010",
"article-title": "The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME–Tox)",
"author": "Sarkadi",
"doi-asserted-by": "crossref",
"first-page": "379",
"journal-title": "Drug Discov. Today",
"key": "ref_70",
"volume": "13",
"year": "2008"
},
{
"DOI": "10.1093/bioinformatics/bty707",
"article-title": "admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "1067",
"journal-title": "Bioinformatics",
"key": "ref_71",
"volume": "35",
"year": "2019"
},
{
"DOI": "10.2174/138161209788682479",
"article-title": "Drug-Like Property Concepts in Pharmaceutical Design",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2184",
"journal-title": "Curr. Pharm. Des.",
"key": "ref_72",
"volume": "15",
"year": "2009"
},
{
"DOI": "10.1517/17460441003605098",
"article-title": "Lipophilicity in drug discovery",
"author": "Waring",
"doi-asserted-by": "crossref",
"first-page": "235",
"journal-title": "Expert. Opin. Drug Discov.",
"key": "ref_73",
"volume": "5",
"year": "2010"
},
{
"DOI": "10.1007/s002280100369",
"article-title": "In vitro prediction of gastrointestinal absorption and bioavailability: An experts’ meeting report",
"author": "Pelkonen",
"doi-asserted-by": "crossref",
"first-page": "621",
"journal-title": "Eur. J. Clin. Pharmacol.",
"key": "ref_74",
"volume": "57",
"year": "2001"
},
{
"DOI": "10.1016/j.ejps.2020.105280",
"article-title": "Impact of gastrointestinal physiology on drug absorption in special populations—An UNGAP review",
"author": "Stillhart",
"doi-asserted-by": "crossref",
"first-page": "105280",
"journal-title": "Eur. J. Pharm. Sci.",
"key": "ref_75",
"volume": "147",
"year": "2020"
},
{
"DOI": "10.7717/peerj.1405",
"article-title": "The apparent permeabilities of Caco-2 cells to marketed drugs: Magnitude, and independence from both biophysical properties and endogenite similarities",
"author": "Kell",
"doi-asserted-by": "crossref",
"first-page": "e1405",
"journal-title": "PeerJ",
"key": "ref_76",
"volume": "3",
"year": "2015"
},
{
"DOI": "10.1023/A:1015937605100",
"article-title": "Caco-2 cell monolayers as a model for drug transport across the intestinal mucosa",
"author": "Hilgers",
"doi-asserted-by": "crossref",
"first-page": "902",
"journal-title": "Pharm. Res.",
"key": "ref_77",
"volume": "7",
"year": "1990"
},
{
"DOI": "10.1007/s13318-019-00602-7",
"article-title": "Evaluation of the Role of P-glycoprotein (P-gp)-Mediated Efflux in the Intestinal Absorption of Common Substrates with Elacridar, a P-gp Inhibitor, in Rats",
"author": "Suzuki",
"doi-asserted-by": "crossref",
"first-page": "385",
"journal-title": "Eur. J. Drug Metab. Pharmacokinet.",
"key": "ref_78",
"volume": "45",
"year": "2020"
},
{
"DOI": "10.3390/pharmaceutics13030388",
"doi-asserted-by": "crossref",
"key": "ref_79",
"unstructured": "Kono, Y., Kawahara, I., Shinozaki, K., Nomura, I., Marutani, H., Yamamoto, A., and Fujita, T. (2021). Characterization of P-Glycoprotein Inhibitors for Evaluating the Effect of P-Glycoprotein on the Intestinal Absorption of Drugs. Pharmaceutics, 13."
},
{
"DOI": "10.1124/dmd.110.032458",
"article-title": "Use of the Øie-Tozer Model in Understanding Mechanisms and Determinants of Drug Distribution",
"author": "Waters",
"doi-asserted-by": "crossref",
"first-page": "1159",
"journal-title": "Drug Metab. Dispos.",
"key": "ref_80",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1016/j.ejps.2010.10.011",
"article-title": "Use of unbound volumes of drug distribution in pharmacokinetic calculations",
"author": "Stepensky",
"doi-asserted-by": "crossref",
"first-page": "91",
"journal-title": "Eur. J. Pharm. Sci.",
"key": "ref_81",
"volume": "42",
"year": "2011"
},
{
"DOI": "10.1007/s11095-016-2086-y",
"article-title": "Drug Distribution Part 2. Predicting Volume of Distribution from Plasma Protein Binding and Membrane Partitioning",
"author": "Korzekwa",
"doi-asserted-by": "crossref",
"first-page": "544",
"journal-title": "Pharm. Res.",
"key": "ref_82",
"volume": "34",
"year": "2017"
},
{
"DOI": "10.1002/cpph.9",
"article-title": "In Vitro Drug Metabolism Using Liver Microsomes",
"author": "Knights",
"doi-asserted-by": "crossref",
"first-page": "7.8.1",
"journal-title": "Curr. Protoc. Pharmacol.",
"key": "ref_83",
"volume": "74",
"year": "2016"
},
{
"DOI": "10.1111/bcp.15990",
"article-title": "Effect of changes in metabolic enzymes and transporters on drug metabolism in the context of liver disease: Impact on pharmacokinetics and drug–drug interactions",
"author": "Armani",
"doi-asserted-by": "crossref",
"first-page": "942",
"journal-title": "Br. J. Clin. Pharmacol.",
"key": "ref_84",
"volume": "90",
"year": "2024"
},
{
"DOI": "10.1016/j.ejps.2010.03.014",
"article-title": "Renal excretion of clofarabine: Assessment of dose-linearity and role of renal transport systems on drug excretion",
"author": "Ajavon",
"doi-asserted-by": "crossref",
"first-page": "209",
"journal-title": "Eur. J. Pharm. Sci.",
"key": "ref_85",
"volume": "40",
"year": "2010"
},
{
"DOI": "10.1124/jpet.117.242552",
"article-title": "Correlation between Apparent Substrate Affinity and OCT2 Transport Turnover",
"author": "Severance",
"doi-asserted-by": "crossref",
"first-page": "405",
"journal-title": "J. Pharmacol. Exp. Ther.",
"key": "ref_86",
"volume": "362",
"year": "2017"
},
{
"DOI": "10.1371/journal.pone.0214862",
"doi-asserted-by": "crossref",
"key": "ref_87",
"unstructured": "Nishizawa, K., Yoda, N., Morokado, F., Komori, H., Nakanishi, T., and Tamai, I. (2019). Changes of drug pharmacokinetics mediated by downregulation of kidney organic cation transporters Mate1 and Oct2 in a rat model of hyperuricemia. PLoS ONE, 14."
},
{
"DOI": "10.1016/j.ejmech.2020.112290",
"article-title": "hERG toxicity assessment: Useful guidelines for drug design",
"author": "Garrido",
"doi-asserted-by": "crossref",
"first-page": "112290",
"journal-title": "European J. Med. Chem.",
"key": "ref_88",
"volume": "195",
"year": "2020"
},
{
"key": "ref_89",
"unstructured": "Wexler, P. (2024). Saccharin. Encyclopedia of Toxicology, Academic Press. [4th ed.]."
},
{
"DOI": "10.1016/S0027-5107(00)00064-6",
"article-title": "The Ames Salmonella/microsome mutagenicity assay",
"author": "Mortelmans",
"doi-asserted-by": "crossref",
"first-page": "29",
"journal-title": "Mutat. Res.-Fundam. Mol. Mech. Mutagen.",
"key": "ref_90",
"volume": "455",
"year": "2000"
},
{
"DOI": "10.1007/s13659-024-00486-4",
"doi-asserted-by": "crossref",
"key": "ref_91",
"unstructured": "Gu, X., Zhang, X., Zhang, X., Wang, X., Sun, W., Zhang, Y., and Hu, Z. (2025). Unveiling the mechanism of action of a novel natural dual inhibitor of SARS-CoV-2 Mpro and PLpro with molecular dynamics simulations. Nat. Prod. Bioprospect., 15."
},
{
"article-title": "Integrated Exploration of Pyranocoumarin Derivatives as Synergistic Inhibitors of Dual-target for Mpro and PLpro Proteins of SARS-CoV-2 through Molecular Docking, ADMET Analysis, and Molecular Dynamics Simulation",
"author": "Imane",
"first-page": "1",
"journal-title": "Curr. Med. Chem.",
"key": "ref_92",
"volume": "31",
"year": "2024"
},
{
"DOI": "10.3389/fmolb.2024.1374364",
"doi-asserted-by": "crossref",
"key": "ref_93",
"unstructured": "Valdés-Albuernes, J.L., Díaz-Pico, E., Alfaro, S., and Caballero, J. (2024). Modeling of noncovalent inhibitors of the papain-like protease (PLpro) from SARS-CoV-2 considering the protein flexibility by using molecular dynamics and cross-docking. Front. Mol. Biosci., 11."
},
{
"DOI": "10.1177/17475198241298547",
"article-title": "Discovery of potential FDA-approved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach",
"author": "Metwaly",
"doi-asserted-by": "crossref",
"first-page": "17475198241298547",
"journal-title": "J. Chem. Res.",
"key": "ref_94",
"volume": "48",
"year": "2024"
},
{
"DOI": "10.1038/s41467-025-57905-4",
"article-title": "A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID",
"author": "Bader",
"doi-asserted-by": "crossref",
"first-page": "2900",
"journal-title": "Nat. Commun.",
"key": "ref_95",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1186/1758-2946-4-17",
"article-title": "Avogadro: An advanced semantic chemical editor, visualization, and analysis platform",
"author": "Hanwell",
"doi-asserted-by": "crossref",
"first-page": "17",
"journal-title": "J. Cheminform.",
"key": "ref_96",
"volume": "4",
"year": "2012"
},
{
"DOI": "10.1038/srep42717",
"doi-asserted-by": "crossref",
"key": "ref_97",
"unstructured": "Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep., 7."
},
{
"DOI": "10.1021/acs.jmedchem.5b00104",
"article-title": "pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures",
"author": "Pires",
"doi-asserted-by": "crossref",
"first-page": "4066",
"journal-title": "J. Med. Chem.",
"key": "ref_98",
"volume": "58",
"year": "2015"
},
{
"DOI": "10.1093/nar/gkab255",
"article-title": "ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties",
"author": "Xiong",
"doi-asserted-by": "crossref",
"first-page": "w5",
"journal-title": "Nucleic Acids Res.",
"key": "ref_99",
"volume": "49",
"year": "2021"
},
{
"DOI": "10.1002/minf.201500040",
"article-title": "Pred-hERG: A Novel web-Accessible Computational Tool for Predicting Cardiac Toxicity",
"author": "Braga",
"doi-asserted-by": "crossref",
"first-page": "698",
"journal-title": "Mol. Inform.",
"key": "ref_100",
"volume": "34",
"year": "2015"
},
{
"DOI": "10.1021/cb500917m",
"article-title": "Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "1456",
"journal-title": "ACS Chem. Biol.",
"key": "ref_101",
"volume": "10",
"year": "2015"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "crossref",
"first-page": "455",
"journal-title": "J. Comput. Chem.",
"key": "ref_102",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1002/jcc.21256",
"article-title": "AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility",
"author": "Morris",
"doi-asserted-by": "crossref",
"first-page": "2785",
"journal-title": "J. Comput. Chem.",
"key": "ref_103",
"volume": "30",
"year": "2009"
},
{
"DOI": "10.1186/1758-2946-3-33",
"article-title": "Open Babel: An open chemical toolbox",
"author": "Banck",
"doi-asserted-by": "crossref",
"first-page": "33",
"journal-title": "J. Cheminform.",
"key": "ref_104",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.3390/ijms25126715",
"doi-asserted-by": "crossref",
"key": "ref_105",
"unstructured": "Bastos, R.S., de Aguiar, C.P.O., Cruz, J.N., Ramos, R.S., Kimani, N.M., de Souza, J.S.N., Chaves, M.H., de Freitas, H.F., Pita, S.S.R., and Santos, C.B.R.d. (2024). Rational Approach toward COVID-19′s Main Protease Inhibitors: A Hierarchical Biochemoinformatics Analysis. Int. J. Mol. Sci., 25."
},
{
"DOI": "10.1016/j.softx.2015.06.001",
"article-title": "GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers",
"author": "Abraham",
"doi-asserted-by": "crossref",
"first-page": "19",
"journal-title": "SoftwareX",
"key": "ref_106",
"volume": "1–2",
"year": "2015"
},
{
"DOI": "10.1038/nmeth.4067",
"article-title": "CHARMM36m: An improved force field for folded and intrinsically disordered proteins",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "71",
"journal-title": "Nat. Methods",
"key": "ref_107",
"volume": "14",
"year": "2017"
},
{
"DOI": "10.1002/jcc.21367",
"article-title": "CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields",
"author": "Vanommeslaeghe",
"doi-asserted-by": "crossref",
"first-page": "671",
"journal-title": "J. Comput. Chem.",
"key": "ref_108",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1021/ci300363c",
"article-title": "Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing",
"author": "Vanommeslaeghe",
"doi-asserted-by": "crossref",
"first-page": "3144",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_109",
"volume": "52",
"year": "2012"
},
{
"DOI": "10.1063/1.464397",
"article-title": "Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems",
"author": "Darden",
"doi-asserted-by": "crossref",
"first-page": "10089",
"journal-title": "J. Chem. Phys.",
"key": "ref_110",
"volume": "98",
"year": "1993"
},
{
"DOI": "10.1016/0021-9991(83)90014-1",
"article-title": "Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations",
"author": "Andersen",
"doi-asserted-by": "crossref",
"first-page": "24",
"journal-title": "J. Comput. Phys.",
"key": "ref_111",
"volume": "52",
"year": "1983"
},
{
"DOI": "10.1063/1.2408420",
"article-title": "Canonical sampling through velocity rescaling",
"author": "Bussi",
"doi-asserted-by": "crossref",
"first-page": "014101",
"journal-title": "J. Chem. Phys.",
"key": "ref_112",
"volume": "126",
"year": "2007"
},
{
"DOI": "10.1063/1.328693",
"article-title": "Polymorphic transitions in single crystals: A new molecular dynamics method",
"author": "Parrinello",
"doi-asserted-by": "crossref",
"first-page": "7182",
"journal-title": "J. Appl. Phys.",
"key": "ref_113",
"volume": "52",
"year": "1981"
},
{
"DOI": "10.1021/ci500020m",
"article-title": "g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations",
"author": "Kumari",
"doi-asserted-by": "crossref",
"first-page": "1951",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_114",
"volume": "54",
"year": "2014"
}
],
"reference-count": 114,
"references-count": 114,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1424-8247/18/6/798"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Structure-Based Design and In-Silico Evaluation of Computationally Proposed Curcumin Derivatives as Potential Inhibitors of the Coronaviral PLpro Enzymes",
"type": "journal-article",
"volume": "18"
}