Discovery of potential FDA-approved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach
et al., Journal of Chemical Research, doi:10.1177/17475198241298547, Nov 2024
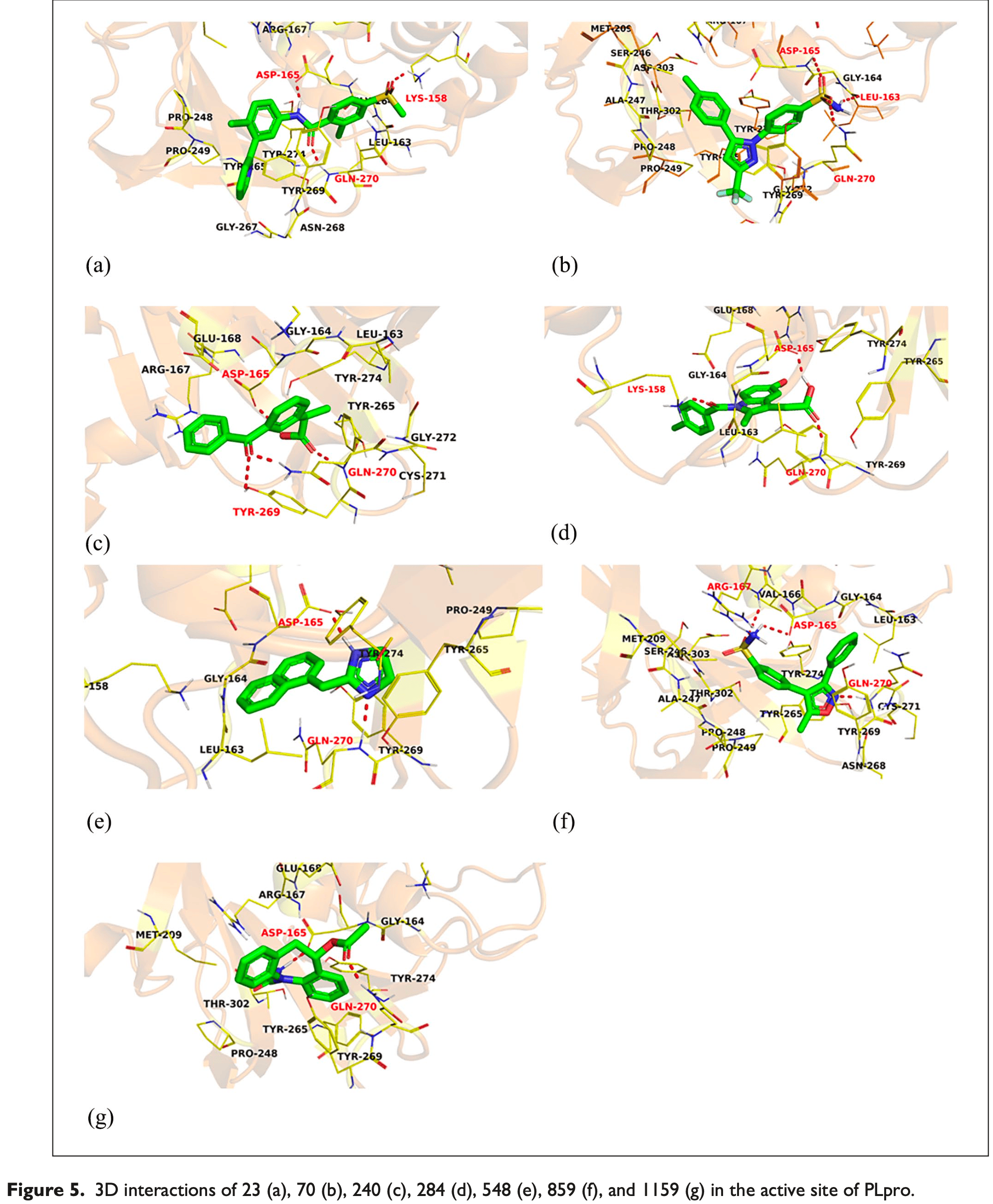
In silico study showing potential inhibition of SARS-CoV-2 papain-like protease (PLpro) by 7 FDA-approved drugs including indomethacin. Authors screened 3,009 drugs and identified indomethacin, vismodegib, celecoxib, ketoprofen, naphazoline, valdecoxib, and eslicarbazepine as showing structural similarity to PLpro's co-crystallized ligand TTT. Molecular docking confirmed these drugs bind PLpro's active site.
7 preclinical studies support the efficacy of indomethacin for COVID-19:
1.
Metwaly et al., Discovery of potential FDA-approved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach, Journal of Chemical Research, doi:10.1177/17475198241298547.
2.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
3.
Chakraborty et al., In-silico screening and in-vitro assay show the antiviral effect of Indomethacin against SARS-CoV-2, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2022.105788.
4.
Tramontozzi et al., Indomethacin inhibits human seasonal coronaviruses at late stages of viral replication in lung cells: Impact on virus-induced COX-2 expression, Journal of Virus Eradication, doi:10.1016/j.jve.2024.100387.
Metwaly et al., 15 Nov 2024, Egypt, peer-reviewed, 7 authors.
Contact: ametwaly@azhar.edu.eg.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Discovery of potential FDA-approved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach
Journal of Chemical Research, doi:10.1177/17475198241298547
Papain-like protease (PLpro) is a crucial enzyme for SARS-CoV-2 replication and immune evasion. Inhibiting PLpro could be a promising strategy to fight against COVID-19. This study aimed to identify potent inhibitors of PLpro among FDAapproved drugs using an in silico approach. The study also aimed to examine and confirm the binding of the selected compounds to the active pocket of PLpro using a multi-phased in silico approach, involving the screening of 3009 FDAapproved drugs to pinpoint the most similar compounds to, TTT, the co-crystallized ligand TTT of PLpro. The selected compounds were subjected to further analysis, including molecular docking, molecular dynamics simulations, MM-GPSA (molecular mechanics generalized Born surface area), and PLIP (Protein-Ligand Interaction Profiler) studies, to examine and confirm their binding to the active pocket of PLpro. Seven candidates (Vismodegib, Celecoxib, Ketoprofen, Indomethacin, Naphazoline, Valdecoxib, and Eslicarbazepine) showed promising in silico activities against the PLpro. The computational analysis confirmed the binding of Celecoxib to the active pocket of PLpro, suggesting its potential in the fight against COVID-19. This study identified seven FDA-approved drugs as potential inhibitors of PLpro, providing a feasible approach for drug repurposing against COVID-19. The results obtained from the in silico approach hold promise, but further in vitro and in vivo studies are warranted to validate the potential of these compounds.
Author contributions
Declaration of conflicting interests The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD Ahmed M Metwaly https://orcid.org/0000-0001-8566-1980
Supplemental material Supplemental material for this article is available online.
References
Altamash, Amhamed, Aparicio, Effect of hydrogen bond donors and acceptors on CO2 absorption by deep eutectic solvents, Processes
Amin, Banerjee, Sing, First structure-activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: an endeavor on COVID-19 drug discovery, Molecul Divers
Baidya, Ghosh, Amin, In silico modeling, identification of crucial molecular fingerprints, and prediction of new possible substrates of human organic cationic transporters 1 and 2, New J Chem
Best, Zhu, Shim, Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles, J Chem Theory Comput
Brooks, Brooks, Mackerell, CHARMM: the biomolecular simulation program, J Comput Chem
Brown, Wobst, A decade of FDA-approved drugs (2010-2019): trends and future directions, J Med Chem
Carvalho, Trincão, Romão, X-ray crystallography in drug discovery, Adv Drug Discov
Chan, Shan, Dahoun, Advancing drug discovery via artificial intelligence, Trend Pharmacol Sci
Chen, Wang, Pang, Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations, Nucleic Acid Res
Chen, Zeng, Wang, Decoding the identification mechanism of an SAM-III riboswitch on ligands through multiple independent Gaussian-accelerated molecular dynamics simulations, J Chem Inform Model
Cheng, Li, Liu, In silico ADMET prediction: recent advances, current challenges and future trends, Curr Med Chem
Chu, He, Wang, In silico design of novel benzohydroxamate-based compounds as inhibitors of histone deacetylase 6 based on 3D-QSAR, molecular docking, and molecular dynamics simulations, New J Chem
Ciociola, Cohen, Kulkarni, How drugs are developed and approved by the FDA: current process and future directions, Am Coll Gastroenterol
Cross, Thompson, Rai, Comparison of several molecular docking programs: pose prediction and virtual screening accuracy, J Chem Inform Model
Duan, Dixon, Lowrie, Analysis and comparison of 2D fingerprints: insights into database screening performance using eight fingerprint methods, J Mol Graph Model
Eissa, Alesawy, Saleh, Ligand and structurebased in silico determination of the most promising SARS-CoV-2 nsp16-nsp10 2′-O-methyltransferase complex inhibitors among 3009 FDA-approved drugs, J Chem
Eissa, Khalifa, Elkaeed, In silico exploration of potential natural inhibitors against SARS-CoV-2 nsp10, Molecules
Eissa, Saleh, St, Multistaged in silico discovery of the best SARS-CoV-2 main protease inhibitors amongst 3009 clinical and FDA-approved compounds, J Chem
Eissa, Yousef, Sami, Exploring the anticancer properties of a new nicotinamide analogue: investigations into in silico analysis, antiproliferative effects, selectivity, VEGFR-2 inhibition, apoptosis induction, and migration suppression, Pathol Res Pract
Elkaeed, Alsfouk, Ibrahim, Computerassisted drug discovery of potential natural inhibitors of the SARS-CoV-2 RNA-dependent RNA polymerase through a multi-phase in silico approach, Antiviral Therapy
Elkaeed, Eissa, Elkady, A multistage in silico study of natural potential inhibitors targeting SARS-CoV-2 main protease, Int J Mol Sci
Elkaeed, Eissa, Saleh, Computer-aided drug discovery of natural antiviral metabolites as potential SARS-CoV-2 helicase inhibitors, J Chem Res
Elkaeed, Elkady, Belal, Multi-phase in silico discovery of potential SARS-CoV-2 RNA-dependent RNA polymerase inhibitors among 3009 clinical and FDAapproved related drugs, J Chem
Elkaeed, Khalifa, Alsfouk, The discovery of potential SARS-CoV-2 natural inhibitors among 4924 African metabolites targeting the papain-like protease: a multi-phase in silico approach, Metabolites
Elkaeed, Metwaly, Alesawy, Discovery of potential SARS-CoV-2 papain-like protease natural inhibitors employing a multi-phase in silico approach, Life
Elkaeed, Youssef, Eissa, Multi-step in silico discovery of natural drugs against COVID-19 targeting main protease, Int J Mol Sci
Engel, Basic overview of chemoinformatics, J Chem Inform Model
Escamilla-Gutiérrez, Ribas-Apaico, Mg, In silico strategies for modeling RNA aptamers and predicting binding sites of their molecular targets, Nucleos Nucleot Nucl Acid
Fan, Fu, Zhang, Progress in molecular docking, Molecules
Grimme, Schreiner, Computational chemistry: the fate of current methods and future challenges, Angewandte Chemie Int Edn
Hassell, An, Bledsoe, Crystallization of protein-ligand complexes, Acta Crystal Sect D: Biol Crystal
Heikamp, Bajorath, How do 2D fingerprints detect structurally diverse active compounds? Revealing compound subset-specific fingerprint features through systematic selection, J Chem Inform Model
Hong, Bang, Anti-inflammatory strategies for schizophrenia: a review of evidence for therapeutic applications and drug repurposing, Clin Psychopharmacol Neurosci
Hospital, Goñi, Orozco, Molecular dynamics simulations: advances and applications, Ann Report Comput Chem
Idris, Yekeen, Alakanse, Computer-aided screening for potential TMPRSS2 inhibitors: a combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches, J Biomol Struct Dynam
Ieritano, Campbell, Hopkins, Predicting differential ion mobility behaviour in silico using machine learning, Analyst
Jo, Kim, Iyer, CHARMM-GUI: a web-based graphical user interface for CHARMM, J Comput Chem
Kaushik, Kumar, Bharadwaj, Ligand-based approach for in-silico drug designing
Klemm, Ebert, Calleja, Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2, EMBO J
Kogej, Engvist, Blomberg, Multifingerprintbased similarity searches for targeted class compound selection, J Chem Inform Model
Konreddy, Lee, Recent drug-repurposing-driven advances in the discovery of novel antibiotics, Curr Med Chem
Lee, Cheng, Swails, CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field, J Chem Theory Comput
Liu, Shi, Zhou, Molecular dynamics simulations and novel drug discovery, Expert Opini Drug Discover
Lu, Li, A new computer model for evaluating the selective binding affinity of phenylalkylamines to T-Type Ca2+ channels, Pharmaceuticals
Maggiora, Vogt, Stumpfe, Molecular similarity in medicinal chemistry: miniperspective, J Med Chem
Metwaly, Alesawy, Alsfouk, Computerassisted drug discovery of potential African anti-SARS-CoV-2 natural products targeting the helicase protein, Nat Prod Commun
Metwaly, Elwan, El-Attar, Structurebased virtual screening, docking, ADMET, molecular dynamics, and MM-PBSA calculations for the discovery of potential natural SARS-CoV-2 helicase inhibitors from traditional Chinese medicine, J Chem
Nowak-Sliwinska, Scapozza, Altaba, Drug repurposing in oncology: compounds, pathways, phenotypes and computational approaches for colorectal cancer, Biochim Biophys Acta Rev Cancer
Opo, Rahman, Ahammad, Structurebased pharmacophore modeling, virtual screening, molecular docking, and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein, Sci Rep
Oprea, Bauman, Bologa, Drug repurposing from an academic perspective, Drug Discover Today: Therapeutic Strat
Oprea, Mestres, Drug repurposing: far beyond new targets for old drugs, AAPS J
Parvathaneni, Kulkarni, Muth, Drug repurposing: a promising tool to accelerate the drug discovery process, Drug Discovery Today
Phillips, Braun, Wang, Scalable molecular dynamics with NAMD, J Comput Chem
Plewczynski, Łaźniewski, Augustyniak, Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database, Mol Inform
Pushpakom, Iorio, Eyers, Drug repurposing: progress, challenges and recommendations, Nature Rev Drug Discover
Ranjan, Devarapalli, Kundu, Isomorphism: molecular similarity to crystal structure similarity in multicomponent forms of analgesic drugs tolfenamic and mefenamic acid, IUCrJ
Rpd, Bank, Protein Data Bank
Sastry, Lowrie, Dixon, Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments, J Chem Inform Model
Schneider, Tanrikulu, Schneider, Self-organizing molecular fingerprints: a ligand-based view on drug-like chemical space and off-target prediction, Futur Med Chem
Shi, Support vector regression-based QSAR models for prediction of antioxidant activity of phenolic compounds, Scientific Reports
Shin, Mukherjee, Grewe, Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity, Nature
Shirley, Sharma, Warren, Drug repurposing of the alcohol abuse medication disulfiram as an anti-parasitic agent, Front Cell Infect Microbiol
Singh, Parida, Lingaraju, Drug repurposing approach to fight COVID-19, Pharmacol Rep
Sleire, Førde, Netland, Drug repurposing in cancer, Pharmacol Res
Spackman, Mckinnon, Fingerprinting intermolecular interactions in molecular crystals, Crystengcomm
Sullivan, Enoch, Ezendam, An adverse outcome pathway for sensitization of the respiratory tract by low-molecular-weight chemicals: building evidence to support the utility of in vitro and in silico methods in a regulatory context, Appl Vitro Toxicol
Taha, Ismail, Ali, Molecular hybridization conceded exceptionally potent quinolinyl-oxadiazole hybrids through phenyl linked thiosemicarbazide antileishmanial scaffolds: in silico validation and SAR studies, Bioorgan Chem
Trivedi, Byrareddy, Drug repurposing approaches to combating viral infections, J Clin Med
Turchi, Cai, An evaluation of in-silico methods for predicting solute partition in multiphase complex fluids: a case study of octanol/water partition coefficient, Chem Eng Sci
Valdés-Tresanco, Valdés-Tresanco, Valiente, gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS, J Chem Theory Comput
Vidal, Serna, Mestres, Ligand-based approaches to in silico pharmacology, Chemoinform Comput Chem Biol
Wan, Tian, Wang, In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using dockingbased 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches, RSC Adv
Westbrook, Burley, How structural biologists and the Protein Data Bank contributed to recent FDA new drug approvals, Structure
Willett, Molecular similarity approaches in chemoinformatics: early history and literature status
Xu, Hagler, Chemoinformatics and drug discovery, Molecules
Zhang, Ren, Ma, Development of an in silico prediction model for chemical-induced urinary tract toxicity by using naïve Bayes classifier, Mol Divers
DOI record:
{
"DOI": "10.1177/17475198241298547",
"ISSN": [
"1747-5198",
"2047-6507"
],
"URL": "http://dx.doi.org/10.1177/17475198241298547",
"abstract": "<jats:p> Papain-like protease (PLpro) is a crucial enzyme for SARS-CoV-2 replication and immune evasion. Inhibiting PLpro could be a promising strategy to fight against COVID-19. This study aimed to identify potent inhibitors of PLpro among FDA-approved drugs using an in silico approach. The study also aimed to examine and confirm the binding of the selected compounds to the active pocket of PLpro using a multi-phased in silico approach, involving the screening of 3009 FDA-approved drugs to pinpoint the most similar compounds to, TTT, the co-crystallized ligand TTT of PLpro. The selected compounds were subjected to further analysis, including molecular docking, molecular dynamics simulations, MM-GPSA (molecular mechanics generalized Born surface area), and PLIP (Protein-Ligand Interaction Profiler) studies, to examine and confirm their binding to the active pocket of PLpro. Seven candidates (Vismodegib, Celecoxib, Ketoprofen, Indomethacin, Naphazoline, Valdecoxib, and Eslicarbazepine) showed promising in silico activities against the PLpro. The computational analysis confirmed the binding of Celecoxib to the active pocket of PLpro, suggesting its potential in the fight against COVID-19. This study identified seven FDA-approved drugs as potential inhibitors of PLpro, providing a feasible approach for drug repurposing against COVID-19. The results obtained from the in silico approach hold promise, but further in vitro and in vivo studies are warranted to validate the potential of these compounds. </jats:p>",
"alternative-id": [
"10.1177/17475198241298547"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0001-8566-1980",
"affiliation": [
{
"name": "Pharmacognosy and Medicinal Plants Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo, Egypt"
}
],
"authenticated-orcid": false,
"family": "Metwaly",
"given": "Ahmed M",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Pharmaceutical Sciences, College of Pharmacy, AlMaarefa University, Riyadh, Saudi Arabia"
}
],
"family": "Elkaeed",
"given": "Eslam B",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pharmaceutical Medicinal Chemistry & Drug Design Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo, Egypt"
}
],
"family": "Khalifa",
"given": "Mohamed M",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pharmaceutical Sciences, College of Pharmacy, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia"
}
],
"family": "Alsfouk",
"given": "Aisha A",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Physics Department, Faculty of Science, Alexandria University, Alexandria, Egypt"
}
],
"family": "Amin",
"given": "Fatma G",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Biophysics Department, Faculty of Science, Cairo University, Giza, Egypt"
}
],
"family": "Ibrahim",
"given": "Ibrahim M",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pharmaceutical Medicinal Chemistry & Drug Design Department, Faculty of Pharmacy (Boys), Al-Azhar University, Cairo, Egypt"
}
],
"family": "Eissa",
"given": "Ibrahim H",
"sequence": "additional"
}
],
"container-title": "Journal of Chemical Research",
"container-title-short": "Journal of Chemical Research",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.sagepub.com"
]
},
"created": {
"date-parts": [
[
2024,
11,
16
]
],
"date-time": "2024-11-16T06:30:28Z",
"timestamp": 1731738628000
},
"deposited": {
"date-parts": [
[
2024,
11,
16
]
],
"date-time": "2024-11-16T06:30:37Z",
"timestamp": 1731738637000
},
"funder": [
{
"DOI": "10.13039/501100004242",
"award": [
"PNURSP2024R116"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100004242",
"id-type": "DOI"
}
],
"name": "Princess Nourah Bint Abdulrahman University"
}
],
"indexed": {
"date-parts": [
[
2024,
11,
16
]
],
"date-time": "2024-11-16T07:40:13Z",
"timestamp": 1731742813169,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "6",
"issued": {
"date-parts": [
[
2024,
11
]
]
},
"journal-issue": {
"issue": "6",
"published-print": {
"date-parts": [
[
2024,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
1
]
],
"date-time": "2024-11-01T00:00:00Z",
"timestamp": 1730419200000
}
}
],
"link": [
{
"URL": "https://journals.sagepub.com/doi/pdf/10.1177/17475198241298547",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.sagepub.com/doi/full-xml/10.1177/17475198241298547",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.sagepub.com/doi/pdf/10.1177/17475198241298547",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "179",
"original-title": [],
"prefix": "10.1177",
"published": {
"date-parts": [
[
2024,
11
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
15
]
]
},
"published-print": {
"date-parts": [
[
2024,
11
]
]
},
"publisher": "SAGE Publications",
"reference": [
{
"DOI": "10.1038/ajg.2013.407",
"doi-asserted-by": "publisher",
"key": "bibr1-17475198241298547"
},
{
"DOI": "10.1021/acs.jmedchem.0c01516",
"doi-asserted-by": "publisher",
"key": "bibr2-17475198241298547"
},
{
"DOI": "10.1038/nrd.2018.168",
"doi-asserted-by": "publisher",
"key": "bibr3-17475198241298547"
},
{
"DOI": "10.1016/j.tips.2019.06.004",
"doi-asserted-by": "publisher",
"key": "bibr4-17475198241298547"
},
{
"DOI": "10.1016/j.drudis.2019.06.014",
"doi-asserted-by": "publisher",
"key": "bibr5-17475198241298547"
},
{
"DOI": "10.1208/s12248-012-9390-1",
"doi-asserted-by": "publisher",
"key": "bibr6-17475198241298547"
},
{
"DOI": "10.1016/j.ddstr.2011.10.002",
"doi-asserted-by": "publisher",
"key": "bibr7-17475198241298547"
},
{
"DOI": "10.1016/j.phrs.2017.07.013",
"doi-asserted-by": "publisher",
"key": "bibr8-17475198241298547"
},
{
"DOI": "10.1016/j.bbcan.2019.04.005",
"doi-asserted-by": "publisher",
"key": "bibr9-17475198241298547"
},
{
"DOI": "10.1007/s43440-020-00155-6",
"doi-asserted-by": "publisher",
"key": "bibr10-17475198241298547"
},
{
"DOI": "10.9758/cpn.2020.18.1.10",
"doi-asserted-by": "publisher",
"key": "bibr11-17475198241298547"
},
{
"DOI": "10.2174/0929867325666180706101404",
"doi-asserted-by": "publisher",
"key": "bibr12-17475198241298547"
},
{
"DOI": "10.3389/fcimb.2021.633194",
"doi-asserted-by": "publisher",
"key": "bibr13-17475198241298547"
},
{
"DOI": "10.3390/jcm9113777",
"doi-asserted-by": "publisher",
"key": "bibr14-17475198241298547"
},
{
"DOI": "10.3390/70800566",
"doi-asserted-by": "publisher",
"key": "bibr15-17475198241298547"
},
{
"DOI": "10.1021/ci600234z",
"doi-asserted-by": "publisher",
"key": "bibr16-17475198241298547"
},
{
"DOI": "10.1016/j.str.2018.11.007",
"doi-asserted-by": "publisher",
"key": "bibr17-17475198241298547"
},
{
"DOI": "10.1016/j.prp.2023.154924",
"doi-asserted-by": "publisher",
"key": "bibr18-17475198241298547"
},
{
"DOI": "10.1002/anie.201709943",
"doi-asserted-by": "publisher",
"key": "bibr19-17475198241298547"
},
{
"DOI": "10.1007/s11030-020-10166-3",
"doi-asserted-by": "publisher",
"key": "bibr20-17475198241298547"
},
{
"DOI": "10.1107/S205225251901604X",
"doi-asserted-by": "publisher",
"key": "bibr21-17475198241298547"
},
{
"DOI": "10.1039/C9NJ05825G",
"doi-asserted-by": "publisher",
"key": "bibr22-17475198241298547"
},
{
"DOI": "10.1038/s41598-021-95859-x",
"doi-asserted-by": "publisher",
"key": "bibr23-17475198241298547"
},
{
"DOI": "10.1080/07391102.2020.1792346",
"doi-asserted-by": "publisher",
"key": "bibr24-17475198241298547"
},
{
"DOI": "10.2174/15680266113139990033",
"doi-asserted-by": "publisher",
"key": "bibr25-17475198241298547"
},
{
"DOI": "10.3390/ph14020141",
"doi-asserted-by": "publisher",
"key": "bibr26-17475198241298547"
},
{
"author": "Fan J",
"first-page": "83",
"journal-title": "Molecules",
"key": "bibr27-17475198241298547",
"volume": "7",
"year": "2019"
},
{
"author": "Hospital A",
"first-page": "37",
"journal-title": "Ann Report Comput Chem",
"key": "bibr28-17475198241298547",
"volume": "2015"
},
{
"DOI": "10.3390/ijms23136912",
"doi-asserted-by": "publisher",
"key": "bibr29-17475198241298547"
},
{
"DOI": "10.3390/ijms23158407",
"doi-asserted-by": "publisher",
"key": "bibr30-17475198241298547"
},
{
"DOI": "10.3390/molecules26206151",
"doi-asserted-by": "publisher",
"key": "bibr31-17475198241298547"
},
{
"DOI": "10.1177/17475198231221253",
"doi-asserted-by": "publisher",
"key": "bibr32-17475198241298547"
},
{
"DOI": "10.3390/life12091407",
"doi-asserted-by": "publisher",
"key": "bibr33-17475198241298547"
},
{
"author": "Elkaeed EB",
"first-page": "530",
"issue": "3",
"journal-title": "J Chem",
"key": "bibr34-17475198241298547",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1155/2024/5084553",
"doi-asserted-by": "publisher",
"key": "bibr35-17475198241298547"
},
{
"author": "Eissa IH",
"first-page": "2287",
"issue": "7",
"journal-title": "J Chem",
"key": "bibr36-17475198241298547",
"volume": "27",
"year": "2022"
},
{
"DOI": "10.1177/13596535231199838",
"doi-asserted-by": "publisher",
"key": "bibr37-17475198241298547"
},
{
"author": "Metwaly AM",
"issue": "4",
"journal-title": "Nat Prod Commun",
"key": "bibr38-17475198241298547",
"volume": "19",
"year": "2024"
},
{
"DOI": "10.3390/metabo12111122",
"doi-asserted-by": "publisher",
"key": "bibr39-17475198241298547"
},
{
"DOI": "10.1155/2022/7270094",
"doi-asserted-by": "publisher",
"key": "bibr40-17475198241298547"
},
{
"DOI": "10.1038/s41586-020-2601-5",
"doi-asserted-by": "publisher",
"key": "bibr41-17475198241298547"
},
{
"DOI": "10.15252/embj.2020106275",
"doi-asserted-by": "publisher",
"key": "bibr42-17475198241298547"
},
{
"key": "bibr43-17475198241298547",
"unstructured": "FDA-approved Drug Library. 2022, https://www.selleckchem.com/screening/fda-approved-drug-library.html (accessed 19 November 2021)."
},
{
"author": "Carvalho AL",
"first-page": "31",
"journal-title": "Adv Drug Discov",
"key": "bibr44-17475198241298547",
"volume": "2010"
},
{
"DOI": "10.1107/S0907444906047020",
"doi-asserted-by": "publisher",
"key": "bibr45-17475198241298547"
},
{
"author": "Vidal D",
"first-page": "489",
"journal-title": "Chemoinform Comput Chem Biol",
"key": "bibr46-17475198241298547",
"volume": "2011"
},
{
"DOI": "10.4155/fmc.09.11",
"doi-asserted-by": "publisher",
"key": "bibr47-17475198241298547"
},
{
"DOI": "10.1039/B203191B",
"doi-asserted-by": "publisher",
"key": "bibr48-17475198241298547"
},
{
"DOI": "10.1039/D0NJ04704J",
"doi-asserted-by": "publisher",
"key": "bibr49-17475198241298547"
},
{
"DOI": "10.1039/D1AN00557J",
"doi-asserted-by": "publisher",
"key": "bibr50-17475198241298547"
},
{
"DOI": "10.1016/j.bioorg.2017.02.005",
"doi-asserted-by": "publisher",
"key": "bibr51-17475198241298547"
},
{
"DOI": "10.1021/ci200275m",
"doi-asserted-by": "publisher",
"key": "bibr52-17475198241298547"
},
{
"DOI": "10.1038/s41598-020-79139-8",
"author": "Opo FADM",
"doi-asserted-by": "crossref",
"first-page": "1",
"issue": "1",
"journal-title": "Sci Rep",
"key": "bibr53-17475198241298547",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.jmgm.2010.05.008",
"doi-asserted-by": "publisher",
"key": "bibr54-17475198241298547"
},
{
"DOI": "10.1021/ci100062n",
"doi-asserted-by": "publisher",
"key": "bibr55-17475198241298547"
},
{
"DOI": "10.1021/ci0504723",
"doi-asserted-by": "publisher",
"key": "bibr56-17475198241298547"
},
{
"author": "Willett P",
"first-page": "67",
"key": "bibr57-17475198241298547",
"volume-title": "Frontiers in molecular design and chemical information science-Herman Skolnik Award Symposium 2015: Jürgen Bajorath (ACS Symposium series)"
},
{
"DOI": "10.1021/jm401411z",
"doi-asserted-by": "publisher",
"key": "bibr58-17475198241298547"
},
{
"DOI": "10.3390/pr8121533",
"doi-asserted-by": "publisher",
"key": "bibr59-17475198241298547"
},
{
"DOI": "10.1039/C8RA06475J",
"doi-asserted-by": "publisher",
"key": "bibr60-17475198241298547"
},
{
"DOI": "10.1016/j.ces.2018.12.003",
"doi-asserted-by": "publisher",
"key": "bibr61-17475198241298547"
},
{
"DOI": "10.1089/aivt.2017.0010",
"doi-asserted-by": "publisher",
"key": "bibr62-17475198241298547"
},
{
"DOI": "10.1080/15257770.2021.1951754",
"doi-asserted-by": "publisher",
"key": "bibr63-17475198241298547"
},
{
"DOI": "10.1007/978-3-319-75732-2_2",
"doi-asserted-by": "publisher",
"key": "bibr64-17475198241298547"
},
{
"DOI": "10.1007/s11030-018-9882-8",
"doi-asserted-by": "publisher",
"key": "bibr65-17475198241298547"
},
{
"DOI": "10.1021/ci900056c",
"doi-asserted-by": "publisher",
"key": "bibr66-17475198241298547"
},
{
"author": "Plewczynski D",
"first-page": "742",
"issue": "4",
"journal-title": "Mol Inform",
"key": "bibr67-17475198241298547",
"volume": "32",
"year": "2011"
},
{
"DOI": "10.1080/17460441.2018.1403419",
"doi-asserted-by": "publisher",
"key": "bibr68-17475198241298547"
},
{
"key": "bibr69-17475198241298547",
"unstructured": "RPD. Bank. Protein Data Bank, 2020, https://www.rcsb.org/structure/4OW0"
},
{
"DOI": "10.1002/jcc.20945",
"doi-asserted-by": "publisher",
"key": "bibr70-17475198241298547"
},
{
"DOI": "10.1002/jcc.21287",
"doi-asserted-by": "publisher",
"key": "bibr71-17475198241298547"
},
{
"DOI": "10.1021/acs.jctc.5b00935",
"doi-asserted-by": "publisher",
"key": "bibr72-17475198241298547"
},
{
"DOI": "10.1021/ct300400x",
"doi-asserted-by": "publisher",
"key": "bibr73-17475198241298547"
},
{
"DOI": "10.1002/jcc.20289",
"doi-asserted-by": "publisher",
"key": "bibr74-17475198241298547"
},
{
"DOI": "10.1021/acs.jctc.1c00645",
"doi-asserted-by": "publisher",
"key": "bibr75-17475198241298547"
},
{
"DOI": "10.1021/acs.jcim.2c00961",
"doi-asserted-by": "publisher",
"key": "bibr76-17475198241298547"
},
{
"DOI": "10.1093/nar/gkz499",
"doi-asserted-by": "publisher",
"key": "bibr77-17475198241298547"
}
],
"reference-count": 77,
"references-count": 77,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.sagepub.com/doi/10.1177/17475198241298547"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Discovery of potential FDA-approved SARS-CoV-2 Papain-like protease inhibitors: A multi-phase in silico approach",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1177/sage-journals-update-policy",
"volume": "48"
}