Exploring SARS-CoV-2 Spike RBD Pockets as Targets for Generic Drugs: A Combined Computational, Biophysical, and Biological Approach
et al., ACS Omega, doi:10.1021/acsomega.5c05175, Aug 2025
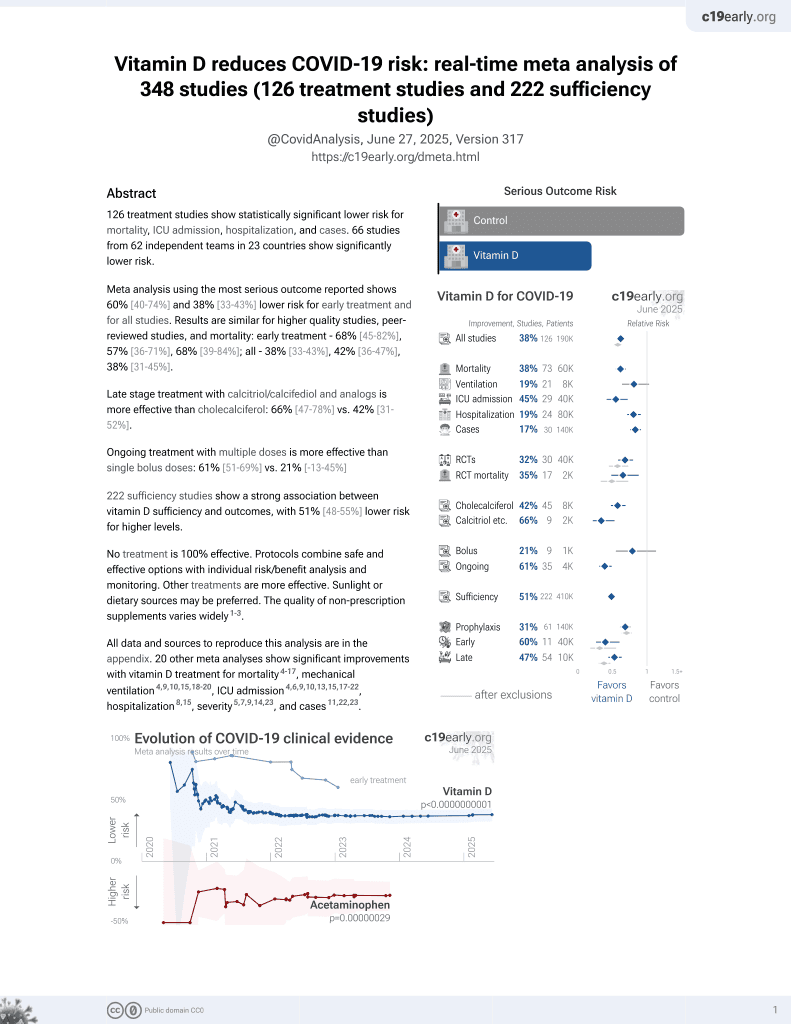
Vitamin D for COVID-19
8th treatment shown to reduce risk in
October 2020, now with p < 0.00000000001 from 136 studies, recognized in 18 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
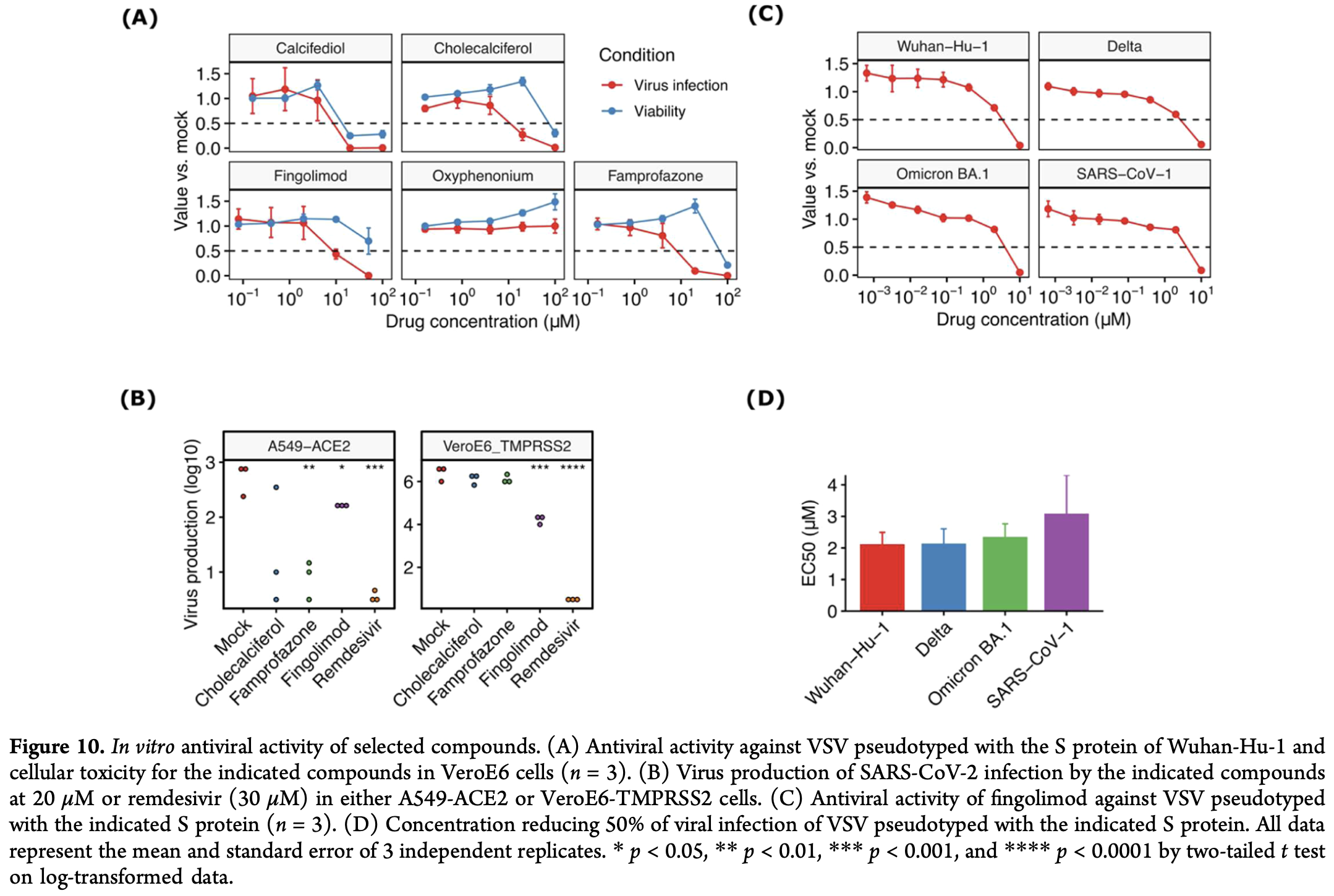
In silico and in vitro study showing that fingolimod inhibits SARS-CoV-2 infection by binding to the spike protein receptor binding domain (RBD). Fingolimod showed the strongest binding affinity to both Wuhan-Hu-1 and Omicron BA.1 variants. Authors also identified other compounds with moderate activity including cholecalciferol, calcifediol, famprofazone, and toremifene.
29 preclinical studies support the efficacy of vitamin D for COVID-19:
Vitamin D has been identified by the European Food Safety Authority (EFSA) as having sufficient evidence for a causal relationship between intake and optimal immune system function27-30.
Vitamin D inhibits SARS-CoV-2 replication in vitro17,24, mitigates lung inflammation, damage, and lethality in mice with an MHV-3 model for β-CoV respiratory infections17,24, reduces SARS-CoV-2 replication in nasal epithelial cells via increased type I interferon expression20, downregulates proinflammatory cytokines IL-1β and TNF-α in SARS-CoV-2 spike protein-stimulated cells16, attenuates nucleocapsid protein-induced hyperinflammation by inactivating the NLRP3 inflammasome through the VDR-BRCC3 signaling pathway21, may be neuroprotective by protecting the blood-brain barrier, reducing neuroinflammation, and via immunomodulatory effects31, may mitigate hyperinflammation and cytokine storm by upregulating TLR10 expression which downregulates proinflammatory cytokines13, downregulates ACE2 and TMPRSS2 in human trophoblasts and minimizes spike protein-induced inflammation19, may minimize cytokine storm by dampening excessive cytokine production2, may suppress viral entry and replication via LL-37 induction11,12, and minimizes platelet aggregation mediated by SARS-CoV-2 spike protein via inhibiting integrin αIIbβ3 outside-in signaling15.
Cholecalciferol and calcifediol directly bind two allosteric pockets on the SARS-CoV-2 Spike RBD, bias the trimer toward a closed state, weaken ACE2 engagement, and reduce viral entry in cell models1.
Calcitriol may destabilize the Spike protein architecture and inhibit IL-17R dimerization, blocking viral entry and mitigating hyperinflammatory cytokine storm32.
Vitamin D improves regulatory immune cell levels and control of proinflammatory cytokines in severe COVID-1933.
Calcifediol inhibits SARS-CoV-2 papain-like protease (PLpro), a critical enzyme for viral replication14.
Symptomatic COVID-19 is associated with a lower frequency of natural killer (NK) cells and vitamin D has been shown to improve NK cell activity34,35.
1.
García-Marín et al., Exploring SARS-CoV-2 Spike RBD Pockets as Targets for Generic Drugs: A Combined Computational, Biophysical, and Biological Approach, ACS Omega, doi:10.1021/acsomega.5c05175.
2.
Alzahrani, A., A new investigation into the molecular mechanism of cholecalciferol towards reducing cytokines storm, Octahedron Drug Research, doi:10.21608/odr.2024.308273.1043.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Morales-Bayuelo et al., New findings on ligand series used as SARS-CoV-2 virus inhibitors within the frameworks of molecular docking, molecular quantum similarity and chemical reactivity indices, F1000Research, doi:10.12688/f1000research.123550.3.
5.
Chellasamy et al., Docking and molecular dynamics studies of human ezrin protein with a modelled SARS-CoV-2 endodomain and their interaction with potential invasion inhibitors, Journal of King Saud University - Science, doi:10.1016/j.jksus.2022.102277.
6.
Pandya et al., Unravelling Vitamin B12 as a potential inhibitor against SARS-CoV-2: A computational approach, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2022.100951.
7.
Mansouri et al., The impact of calcitriol and estradiol on the SARS-CoV-2 biological activity: a molecular modeling approach, Scientific Reports, doi:10.1038/s41598-022-04778-y.
8.
Song et al., Vitamin D3 and its hydroxyderivatives as promising drugs against COVID-19: a computational study, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2021.1964601.
9.
Qayyum et al., Vitamin D and lumisterol novel metabolites can inhibit SARS-CoV-2 replication machinery enzymes, Endocrinology and Metabolism, doi:10.1152/ajpendo.00174.2021.
10.
Al-Mazaideh et al., Vitamin D is a New Promising Inhibitor to the Main Protease (Mpro) of COVID-19 by Molecular Docking, Journal of Pharmaceutical Research International, doi:10.9734/jpri/2021/v33i29B31603.
11.
Roth et al., Vitamin D-inducible antimicrobial peptide LL-37 binds SARS-CoV-2 Spike and accessory proteins ORF7a and ORF8, Frontiers in Cellular and Infection Microbiology, doi:10.3389/fcimb.2025.1671738.
12.
Vercellino et al., Influence of Sex and 1,25α Dihydroxyvitamin D3 on SARS-CoV-2 Infection and Viral Entry, Pathogens, doi:10.3390/pathogens14080765.
13.
Knez et al., TLR10 overexpression modulates immune response in A549 lung epithelial cells challenged with SARS-CoV-2 S and N proteins, Frontiers in Immunology, doi:10.3389/fimmu.2024.1490478.
14.
Chen et al., In Vitro Characterization of Inhibition Function of Calcifediol to the Protease Activity of SARS-COV-2 PLpro, Journal of Medical Virology, doi:10.1002/jmv.70085.
15.
Wang et al., 1,25‐Dihydroxyvitamin D3 attenuates platelet aggregation potentiated by SARS‐CoV‐2 spike protein via inhibiting integrin αIIbβ3 outside‐in signaling, Cell Biochemistry and Function, doi:10.1002/cbf.4039.
16.
Alcalá-Santiago et al., Disentangling the Immunomodulatory Effects of Vitamin D on the SARS-CoV-2 Virus by In Vitro Approaches, The 14th European Nutrition Conference FENS 2023, doi:10.3390/proceedings2023091415.
17.
Campolina-Silva et al., Dietary Vitamin D Mitigates Coronavirus-Induced Lung Inflammation and Damage in Mice, Viruses, doi:10.3390/v15122434.
18.
Moatasim et al., Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E, Microorganisms, doi:10.3390/microorganisms11112777.
19.
Vargas-Castro et al., Calcitriol prevents SARS-CoV spike-induced inflammation in human trophoblasts through downregulating ACE2 and TMPRSS2 expression, The Journal of Steroid Biochemistry and Molecular Biology, doi:10.1016/j.jsbmb.2024.106625.
20.
Sposito et al., Age differential CD13 and interferon expression in airway epithelia affect SARS-CoV-2 infection - effects of vitamin D, Mucosal Immunology, doi:10.1016/j.mucimm.2023.08.002.
21.
Chen (B) et al., Vitamin D3 attenuates SARS‐CoV‐2 nucleocapsid protein‐caused hyperinflammation by inactivating the NLRP3 inflammasome through the VDR‐BRCC3 signaling pathway in vitro and in vivo, MedComm, doi:10.1002/mco2.318.
22.
Rybakovsky et al., Calcitriol modifies tight junctions, improves barrier function, and reduces TNF‐α‐induced barrier leak in the human lung‐derived epithelial cell culture model, 16HBE 14o‐, Physiological Reports, doi:10.14814/phy2.15592.
23.
DiGuilio et al., The multiphasic TNF-α-induced compromise of Calu-3 airway epithelial barrier function, Experimental Lung Research, doi:10.1080/01902148.2023.2193637.
24.
Pickard et al., Discovery of re-purposed drugs that slow SARS-CoV-2 replication in human cells, PLOS Pathogens, doi:10.1371/journal.ppat.1009840.
25.
Mok et al., Calcitriol, the active form of vitamin D, is a promising candidate for COVID-19 prophylaxis, bioRxiv, doi:10.1101/2020.06.21.162396.
26.
Fernandes de Souza et al., Lung Inflammation Induced by Inactivated SARS-CoV-2 in C57BL/6 Female Mice Is Controlled by Intranasal Instillation of Vitamin D, Cells, doi:10.3390/cells12071092.
27.
Galmés et al., Suboptimal Consumption of Relevant Immune System Micronutrients Is Associated with a Worse Impact of COVID-19 in Spanish Populations, Nutrients, doi:10.3390/nu14112254.
28.
Galmés (B) et al., Current State of Evidence: Influence of Nutritional and Nutrigenetic Factors on Immunity in the COVID-19 Pandemic Framework, Nutrients, doi:10.3390/nu12092738.
29.
EFSA, Scientific Opinion on the substantiation of a health claim related to vitamin D and contribution to the normal function of the immune system pursuant to Article 14 of Regulation (EC) No 1924/2006, EFSA Journal, doi:10.2903/j.efsa.2015.4096.
30.
EFSA (B), Scientific Opinion on the substantiation of health claims related to vitamin D and normal function of the immune system and inflammatory response (ID 154, 159), maintenance of normal muscle function (ID 155) and maintenance of normal cardiovascular function (ID 159) pursuant to Article 13(1) of Regulation (E, EFSA Journal, doi:10.2903/j.efsa.2010.1468.
31.
Gotelli et al., Understanding the immune-endocrine effects of vitamin D in SARS-CoV-2 infection: a role in protecting against neurodamage?, Neuroimmunomodulation, doi:10.1159/000533286.
32.
Fadel et al., Targeting asparagine and cysteine in SARS-CoV-2 variants and human pro-inflammatory mediators to alleviate COVID-19 severity; a cross-section and in-silico study, Scientific Reports, doi:10.1038/s41598-025-19359-y.
33.
Saheb Sharif-Askari et al., Increased blood immune regulatory cells in severe COVID-19 with autoantibodies to type I interferons, Scientific Reports, doi:10.1038/s41598-023-43675-w.
García-Marín et al., 25 Aug 2025, peer-reviewed, 10 authors.
Contact: javier.garciamarin@uah.es, smsantamaria@cib.csic.es.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Exploring SARS-CoV-2 Spike RBD Pockets as Targets for Generic Drugs: A Combined Computational, Biophysical, and Biological Approach
ACS Omega, doi:10.1021/acsomega.5c05175
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was a pandemic that killed over 6 million people worldwide, with devastating social and economic impacts still being felt today. Despite the recent and successful development of RNA vaccines, there remains a need for antiviral drugs to treat patients at risk for drug resistance, immunological disorders, or reduced treatment efficacy. In this regard, several computational approaches have been carried out to find small molecules targeting the SARS-CoV-2 Spike S protein, and drug repurposing strategies have been applied to find rapid and accessible candidates for clinical use. In this work, we conduct an exhaustive computational study of the receptor binding domain (RBD) of the spike S protein to identify and characterize druggable pockets and to identify generic drugs as blockers of SARS-CoV-2 entry. The combination of computational screening, biophysical studies in both the RBD (Wuhan-Hu-1 and Omicron BA.1 variants) and Spike protein (Wuhan variant), and the in vitro assays in SARS-CoV-2 Wuhan-Hu-1, Delta, and Omicron BA.1 has led to the identification of generic drugs with S protein binding properties and antiviral activity. Based on in vitro antiviral activity and mechanism of action analysis at the atomic/molecular level, fingolimod exhibited the most promising profile for a possible SARS-CoV-2 antiviral treatment.
■ ASSOCIATED CONTENT
* sı Supporting Information The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.5c05175 .
References
Awad, Abu-Saleh, Sharma, Yadav, Poirier, High-throughput virtual screening of drug databanks for potential inhibitors of SARS-CoV-2 spike glycoprotein, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1835721
Baum, Ajithdoss, Copin, Zhou, Lanza et al., REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters, Science, doi:10.1126/science.abe2402
Behloul, Baha, Guo, Yang, Shi et al., In silico identification of strong binders of the SARS-CoV-2 receptorbinding domain, Eur. J. Pharmacol, doi:10.1016/j.ejphar.2020.173701
Berendsen, Postma, Van Gunsteren, Dinola, Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys, doi:10.1063/1.448118
Cao, Goreshnik, Coventry, Case, Miller et al., De novo design of picomolar SARS-CoV-2 miniprotein inhibitors, Science, doi:10.1126/science.abd9909
Casalino, Gaieb, Goldsmith, Hjorth, Dommer et al., Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein, ACS Cent. Sci, doi:10.1021/acscentsci.0c01056?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Case, None
Castillo, Costa, Barrios, Díaz, Miranda et al., Effect of calcifediol treatment and best available therapy versus best available therapy on intensive care unit admission and mortality among patients hospitalized for COVID-19: A pilot randomized clinical study, J. Steroid Biochem. Mol. Biol, doi:10.1016/j.jsbmb.2020.105751
De Vivo, Masetti, Bottegoni, Cavalli, Role of Molecular Dynamics and Related Methods in Drug Discovery, J. Med. Chem, doi:10.1021/acs.jmedchem.5b01684?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Deganutti, Prischi, Reynolds, Supervised molecular dynamics for exploring the druggability of the SARS-CoV-2 spike protein, J. Comput.-Aided Mol. Des, doi:10.1007/s10822-020-00356-4
Dong, Dai, Wang, Zhang, Zeng et al., The way of SARS-CoV-2 vaccine development: success and challenges, Signal Transduction Targeted Ther, doi:10.1038/s41392-021-00796-w
Eisenberg, Schwarz, Komaromy, Wall, Analysis of membrane and surface protein sequences with the hydrophobic moment plot, J. Mol. Biol, doi:10.1016/0022-2836(84)90309-7
Friesner, Banks, Murphy, Halgren, Klicic et al., Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J. Med. Chem, doi:10.1021/jm0306430?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
García-Iriepa, Hognon, Francés-Monerris, Iriepa, Miclot et al., Thermodynamics of the Interaction between the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus-2 and the Receptor of Human Angiotensin-Converting Enzyme 2. Effects of Possible Ligands, J. Phys. Chem. Lett, doi:10.1021/acs.jpclett.0c02203?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Gargantilla, Francés, Adhav, Forcada-Nadal, Martínez-Gualda et al., C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein, J. Med. Chem, doi:10.1021/acs.jmedchem.3c00576?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Ginex, Marco-Marín, Wieczór, Mata, Krieger et al., The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation, PLoS Pathog, doi:10.1371/journal.ppat.1010995
Grant, Lahore, Mcdonnell, Baggerly, French et al., Evidence that Vitamin D Supplementation Could Reduce Risk of Influenza and COVID-19 Infections and Deaths, Nutrients, doi:10.3390/nu12040988
Grant, Montgomery, Ito, Woods, Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition, Sci. Rep, doi:10.1038/s41598-020-71748-7
Greenwood, Calkins, Sullivan, Shelley, Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution, J. Comput.-Aided Mol. Des, doi:10.1007/s10822-010-9349-1
Gulotta, Lombino, Perricone, De Simone, Mekni et al., Targeting SARS-CoV-2 RBD Interface: a Supervised Computational Data-Driven Approach to Identify Potential Modulators, ChemMedChem, doi:10.1002/cmdc.202000259
Halgren, Identifying and characterizing binding sites and assessing druggability, J. Chem. Inf. Model, doi:10.1021/ci800324m?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Halgren, Murphy, Friesner, Beard, Frye et al., Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening, J. Med. Chem, doi:10.1021/jm030644s?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Halgren, New method for fast and accurate binding-site identification and analysis, Chem. Biol. Drug Des, doi:10.1111/j.1747-0285.2007.00483.x
Harris, Millman, Van Der Walt, Gommers, Virtanen et al., Array programming with NumPy, Nature, doi:10.1038/s41586-020-2649-2
Harvey, Carabelli, Jackson, Gupta, Thomson et al., COVID-19 Genomics UK
Holmes, Structural biology. Adaptation of SARS coronavirus to humans, Science, doi:10.1126/science.1118817
Hsieh, Goldsmith, Schaub, Divenere, Kuo et al., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes, Science, doi:10.1126/science.abd0826
Humphrey, Dalke, Schulten, Vmd, Visual molecular dynamics, J. Mol. Graphics, doi:10.1016/0263-7855(96)00018-5
Hunter, Matplotlib: A 2D Graphics Environment, Comput. Sci. Eng, doi:10.1109/MCSE.2007.55
Jakalian, Jack, Bayly, Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation, J. Comput. Chem, doi:10.1002/jcc.10128
Jo, Kim, Iyer, Im, CHARMM-GUI: a webbased graphical user interface for CHARMM, J. Comput. Chem, doi:10.1002/jcc.20945
Jorgensen, Chandrasekhar, Madura, Impey, Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys, doi:10.1063/1.445869
Kirschner, Yongye, Tschampel, González-Outeirinõ, Daniels et al., GLYCAM06: a generalizable biomolecular force field, Carbohydrates. J. Comput. Chem, doi:10.1002/jcc.20820
Lan, Ge, Yu, Shan, Zhou et al., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature, doi:10.1038/s41586-020-2180-5
Li, Chi, Su, Ferrall, Hung et al., Coronavirus vaccine development: from SARS and MERS to COVID-19, J. Biomed. Sci, doi:10.1186/s12929-020-00695-2
Li, Li, Farzan, Harrison, Structure of SARS coronavirus spike receptor-binding domain complexed with receptor, Science, doi:10.1126/science.1116480
Liu, Wei, Kappler, Marrack, Zhang, SARS-CoV-2 Variants of Concern and Variants of Interest Receptor Binding Domain Mutations and Virus Infectivity, Front. Immunol, doi:10.3389/fimmu.2022.825256
Maier, Martinez, Kasavajhala, Wickstrom, Hauser et al., ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput, doi:10.1021/acs.jctc.5b00255?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Mcgibbon, Beauchamp, Harrigan, Klein, Swails et al., A Modern Open Library for the Analysis of Molecular Dynamics Trajectories, Biophys. J, doi:10.1016/j.bpj.2015.08.015
Michaud-Agrawal, Denning, Woolf, Beckstein, MDAnalysis: a toolkit for the analysis of molecular dynamics simulations, J. Comput. Chem, doi:10.1002/jcc.21787
Nogues, Ovejero, Pineda-Moncusí, Bouillon, Arenas et al., Calcifediol Treatment and COVID-19-Related Outcomes, J. Clin. Endocrinol. Metab, doi:10.1210/clinem/dgab405
Nooruzzaman, Johnson, Rani, Finkelsztein, Caserta et al., Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections, Nat. Commun, doi:10.1038/s41467-024-51924-3
Osuchowski, Winkler, Skirecki, Cajander, Shankar-Hari et al., The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity, Lancet Respir. Med, doi:10.1016/S2213-2600(21)00218-6
Pan, Peto, Restrepo, WHO Solidarity Trial Consortium Remdesivir and three other drugs for hospitalised patients with COVID-19: final results of the WHO Solidarity randomised trial and updated meta-analyses, Lancet, doi:10.1016/S0140-6736(22)00519-0?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Park, Lee, Qi, Kern, Lee et al., CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates, Glycobiology, doi:10.1093/glycob/cwz003
Peacock, Robertson, SARS-CoV-2 variants, spike mutations and immune escape, Nat. Rev. Microbiol, doi:10.1038/s41579-021-00573-0
Perola, Herman, Weiss, Development of a rule-based method for the assessment of protein druggability, J. Chem. Inf. Model, doi:10.1021/ci200613b?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Pettersen, Goddard, Huang, Meng, Couch et al., UCSF ChimeraX: Structure visualization for researchers, educators, and developers, Protein Sci, doi:10.1002/pro.3943
Pomplun, Jbara, Quartararo, Zhang, Brown et al., De Novo Discovery of High-Affinity Peptide Binders for the SARS-CoV-2 Spike Protein, ACS Cent. Sci, doi:10.1021/acscentsci.0c01309?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Puhl, Fritch, Lane, Tse, Yount et al., Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms, ACS Omega, doi:10.1021/acsomega.0c05996?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Pérez-Regidor, Guzmán-Caldentey, Oberhauser, Punzón, Balogh et al., Small Molecules as Toll-like Receptor 4 Modulators Drug and In-House Computational Repurposing, Biomedicines, doi:10.3390/biomedicines10092326
Rarey, Kramer, Lengauer, Klebe, A fast flexible docking method using an incremental construction algorithm, J. Mol. Biol, doi:10.1006/jmbi.1996.0477
Salomon-Ferrer, Case, Walker, An overview of the Amber biomolecular simulation package, WIREs Comput. Mol. Sci, doi:10.1002/wcms.1121
Schneider, Lange, Hindle, Klein, Rarey, A consistent description of HYdrogen bond and DEhydration energies in protein-ligand complexes: methods behind the HYDE scoring function, J. Comput.-Aided Mol. Des, doi:10.1007/s10822-012-9626-2
Shajahan, Supekar, Gleinich, Azadi, Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2, Glycobiology, doi:10.1093/glycob/cwaa042
Shang, Ye, Shi, Wan, Luo et al., Structural basis of receptor recognition by SARS-CoV-2, Nature, doi:10.1038/s41586-020-2179-y
Singh, Villoutreix, Resources and computational strategies to advance small molecule SARS-CoV-2 discovery: Lessons from the pandemic and preparing for future health crises, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2021.04.059
Sztain, Ahn, Bogetti, Casalino, Goldsmith et al., A glycan gate controls opening of the SARS-CoV-2 spike protein, Nat. Chem, doi:10.1038/s41557-021-00758-3
Teymouri, Pourbayram Kaleybar, Hejazian, Hejazian, Ansarin et al., The effect of Fingolimod on patients with moderate to severe COVID-19, Pharmacol. Res. Perspect, doi:10.1002/prp2.1039
Toelzer, Gupta, Yadav, Borucu, Davidson et al., Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein, Science, doi:10.1126/science.abd3255
Touret, Gilles, Barral, Nougairede, Van Helden et al., In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication, Sci. Rep, doi:10.1038/s41598-020-70143-6
Trigueiro-Louro, Correia, Santos, Guedes, Brito et al., To hit or not to hit: Large-scale sequence analysis and structure characterization of influenza A NS1 unlocks new antiviral target potential, Virology, doi:10.1016/j.virol.2019.04.009
Van Gunsteren, Berendsen, Algorithms for macromolecular dynamics and constraint dynamics, Mol. Phys, doi:10.1080/00268977700102571
Vedadi, Niesen, Allali-Hassani, Fedorov, Finerty et al., Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination, Proc. Natl. Acad. Sci. U.S.A, doi:10.1073/pnas.0605224103
Vitiello, Zovi, Rezza, New emerging SARS-CoV-2 variants and antiviral agents, Drug Resist. Updat, doi:10.1016/j.drup.2023.100986
Volkamer, Kuhn, Grombacher, Rippmann, Rarey, Combining global and local measures for structure-based druggability predictions, J. Chem. Inf. Model, doi:10.1021/ci200454v?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Wang, Wolf, Caldwell, Kollman, Case, Development and testing of a general amber force field, J. Comput. Chem, doi:10.1002/jcc.20035
Wang, Wu, Yao, Ge, Zhu et al., Discovery of potential small molecular SARS-CoV-2 entry blockers targeting the spike protein, Acta Pharmacol. Sin, doi:10.1038/s41401-021-00735-z
Watanabe, Allen, Wrapp, Mclellan, Crispin, Site-specific glycan analysis of the SARS-CoV-2 spike, Science, doi:10.1126/science.abb9983
Wienken, Baaske, Rothbauer, Braun, Duhr, Protein-binding assays in biological liquids using microscale thermophoresis, Nat. Commun, doi:10.1038/ncomms1093
Woo, Park, Choi, Park, Tanveer et al., Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane, J. Phys. Chem. B, doi:10.1021/acs.jpcb.0c04553?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Wu, Yuan, Liu, Lee, Zhu et al., An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain, Cell. Rep, doi:10.1016/j.celrep.2020.108274
Xiong, Xiang, Huang, Liu, Wang et al., Structure-Based Virtual Screening and Identification of Potential Inhibitors of SARS-CoV-2 S-RBD and ACE2 Interaction, Front. Chem, doi:10.3389/fchem.2021.740702
Zhao, Praissman, Grant, Cai, Xiao et al., Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor, Cell Host Microbe, doi:10.1016/j.chom.2020.08.004
DOI record:
{
"DOI": "10.1021/acsomega.5c05175",
"ISSN": [
"2470-1343",
"2470-1343"
],
"URL": "http://dx.doi.org/10.1021/acsomega.5c05175",
"alternative-id": [
"10.1021/acsomega.5c05175"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-5883-4783",
"affiliation": [
{
"name": "Centro de Investigaciones Biológicas Margarita Salas (CIB), CSIC",
"place": [
"Madrid, Spain"
]
}
],
"authenticated-orcid": true,
"family": "García-Marín",
"given": "Javier",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute for Integrative Systems Biology (I2SysBio), UV-CSIC",
"place": [
"Paterna, Spain"
]
}
],
"family": "Francés-Gómez",
"given": "Clara",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0179-4044",
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
},
{
"name": "Group 739 at the IBV-CSIC of the Centro de Investigación Biomédica en Red en Enfermedades Raras",
"place": [
"Madrid, Spain"
]
},
{
"name": "Instituto de Salud Carlos III (CIBERER-ISCIII)",
"place": [
"Madrid, Spain"
]
}
],
"authenticated-orcid": true,
"family": "Forcada-Nadal",
"given": "Alicia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
}
],
"family": "Adhav",
"given": "Anmol",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
}
],
"family": "Marco-Marín",
"given": "Clara",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
},
{
"name": "Group 739 at the IBV-CSIC of the Centro de Investigación Biomédica en Red en Enfermedades Raras",
"place": [
"Madrid, Spain"
]
},
{
"name": "Instituto de Salud Carlos III (CIBERER-ISCIII)",
"place": [
"Madrid, Spain"
]
}
],
"family": "Rubio",
"given": "Vicente",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1334-5273",
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
},
{
"name": "Group 739 at the IBV-CSIC of the Centro de Investigación Biomédica en Red en Enfermedades Raras",
"place": [
"Madrid, Spain"
]
},
{
"name": "Instituto de Salud Carlos III (CIBERER-ISCIII)",
"place": [
"Madrid, Spain"
]
}
],
"authenticated-orcid": true,
"family": "Marina",
"given": "Alberto",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5304-1795",
"affiliation": [
{
"name": "Instituto de Biomedicina de Valencia (IBV), CSIC",
"place": [
"Valencia, Spain"
]
},
{
"name": "Group 739 at the IBV-CSIC of the Centro de Investigación Biomédica en Red en Enfermedades Raras",
"place": [
"Madrid, Spain"
]
},
{
"name": "Instituto de Salud Carlos III (CIBERER-ISCIII)",
"place": [
"Madrid, Spain"
]
}
],
"authenticated-orcid": true,
"family": "Llácer",
"given": "José-Luis",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute for Integrative Systems Biology (I2SysBio), UV-CSIC",
"place": [
"Paterna, Spain"
]
}
],
"family": "Geller",
"given": "Ron",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7679-0155",
"affiliation": [
{
"name": "Centro de Investigaciones Biológicas Margarita Salas (CIB), CSIC",
"place": [
"Madrid, Spain"
]
}
],
"authenticated-orcid": true,
"family": "Martín-Santamaría",
"given": "Sonsoles",
"sequence": "additional"
}
],
"container-title": "ACS Omega",
"container-title-short": "ACS Omega",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
8,
25
]
],
"date-time": "2025-08-25T11:35:06Z",
"timestamp": 1756121706000
},
"deposited": {
"date-parts": [
[
2025,
9,
9
]
],
"date-time": "2025-09-09T09:27:56Z",
"timestamp": 1757410076000
},
"funder": [
{
"DOI": "10.13039/100014440",
"award": [
"PID2020-113588RB-I00",
"PID2020-120322RB-C21",
"PID2023-152271NB-I00"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100014440",
"id-type": "DOI"
}
],
"name": "Ministerio de Ciencia, Innovaci?n y Universidades"
},
{
"DOI": "10.13039/100031478",
"award": [
"CSIC-COV19-082"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100031478",
"id-type": "DOI"
}
],
"name": "NextGenerationEU"
}
],
"indexed": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T20:50:15Z",
"timestamp": 1757623815750,
"version": "3.44.0"
},
"is-referenced-by-count": 0,
"issue": "35",
"issued": {
"date-parts": [
[
2025,
8,
25
]
]
},
"journal-issue": {
"issue": "35",
"published-print": {
"date-parts": [
[
2025,
9,
9
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
8,
25
]
],
"date-time": "2025-08-25T00:00:00Z",
"timestamp": 1756080000000
}
}
],
"link": [
{
"URL": "https://pubs.acs.org/doi/pdf/10.1021/acsomega.5c05175",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "unspecified"
},
{
"URL": "https://pubs.acs.org/doi/pdf/10.1021/acsomega.5c05175",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "316",
"original-title": [],
"page": "40190-40207",
"prefix": "10.1021",
"published": {
"date-parts": [
[
2025,
8,
25
]
]
},
"published-online": {
"date-parts": [
[
2025,
8,
25
]
]
},
"published-print": {
"date-parts": [
[
2025,
9,
9
]
]
},
"publisher": "American Chemical Society (ACS)",
"reference": [
{
"DOI": "10.1016/S2213-2600(21)00218-6",
"doi-asserted-by": "publisher",
"key": "ref1/cit1"
},
{
"DOI": "10.1038/s41392-021-00796-w",
"doi-asserted-by": "publisher",
"key": "ref2/cit2"
},
{
"DOI": "10.1186/s12929-020-00695-2",
"doi-asserted-by": "publisher",
"key": "ref3/cit3"
},
{
"DOI": "10.1016/S0140-6736(22)00519-0",
"doi-asserted-by": "publisher",
"key": "ref4/cit4"
},
{
"DOI": "10.1016/j.drup.2023.100986",
"doi-asserted-by": "publisher",
"key": "ref5/cit5"
},
{
"DOI": "10.1038/s41467-024-51924-3",
"doi-asserted-by": "publisher",
"key": "ref6/cit6"
},
{
"DOI": "10.1126/science.1116480",
"doi-asserted-by": "publisher",
"key": "ref7/cit7"
},
{
"DOI": "10.1126/science.1118817",
"doi-asserted-by": "publisher",
"key": "ref8/cit8"
},
{
"DOI": "10.1038/s41586-020-2180-5",
"doi-asserted-by": "publisher",
"key": "ref9/cit9"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"doi-asserted-by": "publisher",
"key": "ref10/cit10"
},
{
"DOI": "10.1021/acs.jmedchem.5b01684",
"doi-asserted-by": "publisher",
"key": "ref11/cit11"
},
{
"DOI": "10.1021/acscentsci.0c01056",
"doi-asserted-by": "publisher",
"key": "ref12/cit12"
},
{
"DOI": "10.1126/science.abe2402",
"doi-asserted-by": "publisher",
"key": "ref13/cit13"
},
{
"DOI": "10.1016/j.celrep.2020.108274",
"doi-asserted-by": "publisher",
"key": "ref14/cit14"
},
{
"DOI": "10.1126/science.abd3255",
"doi-asserted-by": "publisher",
"key": "ref15/cit15"
},
{
"DOI": "10.1021/acs.jpclett.0c02203",
"doi-asserted-by": "publisher",
"key": "ref16/cit16"
},
{
"DOI": "10.1080/07391102.2020.1835721",
"doi-asserted-by": "publisher",
"key": "ref17/cit17"
},
{
"DOI": "10.1002/cmdc.202000259",
"doi-asserted-by": "publisher",
"key": "ref18/cit18"
},
{
"DOI": "10.1007/s10822-020-00356-4",
"doi-asserted-by": "publisher",
"key": "ref19/cit19"
},
{
"DOI": "10.1016/j.csbj.2021.04.059",
"doi-asserted-by": "publisher",
"key": "ref20/cit20"
},
{
"DOI": "10.1038/s41401-021-00735-z",
"doi-asserted-by": "publisher",
"key": "ref21/cit21"
},
{
"DOI": "10.1126/science.abd9909",
"doi-asserted-by": "publisher",
"key": "ref22/cit22"
},
{
"DOI": "10.1021/acscentsci.0c01309",
"doi-asserted-by": "publisher",
"key": "ref23/cit23"
},
{
"DOI": "10.1021/acsomega.0c05996",
"doi-asserted-by": "publisher",
"key": "ref24/cit24"
},
{
"DOI": "10.1021/ci800324m",
"doi-asserted-by": "publisher",
"key": "ref25/cit25"
},
{
"DOI": "10.1021/ci200454v",
"doi-asserted-by": "publisher",
"key": "ref26/cit26"
},
{
"DOI": "10.1021/ci200613b",
"doi-asserted-by": "publisher",
"key": "ref27/cit27"
},
{
"DOI": "10.1093/glycob/cwaa042",
"doi-asserted-by": "publisher",
"key": "ref28/cit28"
},
{
"DOI": "10.1016/j.chom.2020.08.004",
"doi-asserted-by": "publisher",
"key": "ref29/cit29"
},
{
"DOI": "10.3390/biomedicines10092326",
"doi-asserted-by": "publisher",
"key": "ref30/cit30"
},
{
"DOI": "10.1016/j.ejphar.2020.173701",
"doi-asserted-by": "publisher",
"key": "ref31/cit31"
},
{
"DOI": "10.3389/fchem.2021.740702",
"doi-asserted-by": "publisher",
"key": "ref32/cit32"
},
{
"DOI": "10.1038/s41598-020-70143-6",
"doi-asserted-by": "publisher",
"key": "ref33/cit33"
},
{
"DOI": "10.1126/science.abb9983",
"doi-asserted-by": "publisher",
"key": "ref34/cit34"
},
{
"DOI": "10.1016/j.jsbmb.2020.105751",
"doi-asserted-by": "publisher",
"key": "ref35/cit35"
},
{
"DOI": "10.1210/clinem/dgab405",
"doi-asserted-by": "publisher",
"key": "ref36/cit36"
},
{
"DOI": "10.3390/nu12040988",
"doi-asserted-by": "publisher",
"key": "ref37/cit37"
},
{
"DOI": "10.1038/s41557-021-00758-3",
"doi-asserted-by": "publisher",
"key": "ref38/cit38"
},
{
"DOI": "10.1038/s41598-020-71748-7",
"doi-asserted-by": "publisher",
"key": "ref39/cit39"
},
{
"key": "ref40/cit40",
"unstructured": "AnonymousCOVID-19 Molecular Structure and Therapeutics\nHub. https://covid.molssi.org (accessed Feb 21, 2025)."
},
{
"key": "ref41/cit41",
"unstructured": "AnonymousCOVID-19\nMolecular Structure Archive. http://www.charmm-gui.org/?doc=archive&lib=covid19 (accessed Feb 15, 2025)."
},
{
"DOI": "10.1038/s41579-021-00573-0",
"doi-asserted-by": "publisher",
"key": "ref42/cit42"
},
{
"key": "ref43/cit43",
"unstructured": "WHO Tracking SARS-CoV-2 Variants; World Health Organization. https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed April 11, 2022)."
},
{
"DOI": "10.3389/fimmu.2022.825256",
"doi-asserted-by": "publisher",
"key": "ref44/cit44"
},
{
"DOI": "10.1016/0022-2836(84)90309-7",
"doi-asserted-by": "publisher",
"key": "ref45/cit45"
},
{
"DOI": "10.1002/prp2.1039",
"doi-asserted-by": "publisher",
"key": "ref46/cit46"
},
{
"DOI": "10.1093/glycob/cwz003",
"doi-asserted-by": "publisher",
"key": "ref47/cit47"
},
{
"DOI": "10.1002/jcc.20945",
"doi-asserted-by": "publisher",
"key": "ref48/cit48"
},
{
"DOI": "10.1021/acs.jpcb.0c04553",
"doi-asserted-by": "publisher",
"key": "ref49/cit49"
},
{
"DOI": "10.1111/j.1747-0285.2007.00483.x",
"doi-asserted-by": "publisher",
"key": "ref50/cit50"
},
{
"DOI": "10.1016/j.virol.2019.04.009",
"doi-asserted-by": "publisher",
"key": "ref51/cit51"
},
{
"DOI": "10.1007/s10822-010-9349-1",
"doi-asserted-by": "publisher",
"key": "ref52/cit52"
},
{
"DOI": "10.1021/jm0306430",
"doi-asserted-by": "publisher",
"key": "ref53/cit53"
},
{
"DOI": "10.1021/jm030644s",
"doi-asserted-by": "publisher",
"key": "ref54/cit54"
},
{
"DOI": "10.1006/jmbi.1996.0477",
"doi-asserted-by": "publisher",
"key": "ref55/cit55"
},
{
"DOI": "10.1007/s10822-012-9626-2",
"doi-asserted-by": "publisher",
"key": "ref56/cit56"
},
{
"DOI": "10.1021/acs.jctc.5b00255",
"doi-asserted-by": "publisher",
"key": "ref57/cit57"
},
{
"DOI": "10.1002/jcc.20820",
"doi-asserted-by": "publisher",
"key": "ref58/cit58"
},
{
"DOI": "10.1002/jcc.20035",
"doi-asserted-by": "publisher",
"key": "ref59/cit59"
},
{
"DOI": "10.1002/jcc.10128",
"doi-asserted-by": "publisher",
"key": "ref60/cit60"
},
{
"DOI": "10.1002/wcms.1121",
"doi-asserted-by": "publisher",
"key": "ref61/cit61"
},
{
"DOI": "10.1063/1.445869",
"doi-asserted-by": "publisher",
"key": "ref62/cit62"
},
{
"DOI": "10.1080/00268977700102571",
"doi-asserted-by": "publisher",
"key": "ref63/cit63"
},
{
"DOI": "10.1063/1.448118",
"doi-asserted-by": "publisher",
"key": "ref64/cit64"
},
{
"key": "ref100/cit100",
"unstructured": "Case, D. A., et al. AMBER 2018, University of California, San Francisco, https://ambermd.org/doc12/Amber18.pdf, 2018."
},
{
"DOI": "10.1016/j.bpj.2015.08.015",
"doi-asserted-by": "publisher",
"key": "ref65/cit65"
},
{
"DOI": "10.1002/jcc.21787",
"doi-asserted-by": "publisher",
"key": "ref66/cit66"
},
{
"DOI": "10.1038/s41586-020-2649-2",
"doi-asserted-by": "publisher",
"key": "ref67/cit67"
},
{
"DOI": "10.1109/MCSE.2007.55",
"doi-asserted-by": "publisher",
"key": "ref68/cit68"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"doi-asserted-by": "publisher",
"key": "ref69/cit69"
},
{
"DOI": "10.1002/pro.3943",
"doi-asserted-by": "publisher",
"key": "ref70/cit70"
},
{
"DOI": "10.1021/acs.jmedchem.3c00576",
"doi-asserted-by": "publisher",
"key": "ref71/cit71"
},
{
"DOI": "10.1371/journal.ppat.1010995",
"doi-asserted-by": "publisher",
"key": "ref72/cit72"
},
{
"DOI": "10.1126/science.abd0826",
"doi-asserted-by": "publisher",
"key": "ref73/cit73"
},
{
"DOI": "10.1073/pnas.0605224103",
"doi-asserted-by": "publisher",
"key": "ref74/cit74"
},
{
"DOI": "10.1038/ncomms1093",
"doi-asserted-by": "publisher",
"key": "ref75/cit75"
}
],
"reference-count": 76,
"references-count": 76,
"relation": {},
"resource": {
"primary": {
"URL": "https://pubs.acs.org/doi/10.1021/acsomega.5c05175"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Exploring SARS-CoV-2 Spike RBD Pockets as Targets for Generic Drugs: A Combined Computational, Biophysical, and Biological Approach",
"type": "journal-article",
"volume": "10"
}