Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections
et al., Nature Communications, doi:10.1038/s41467-024-51924-3, Sep 2024
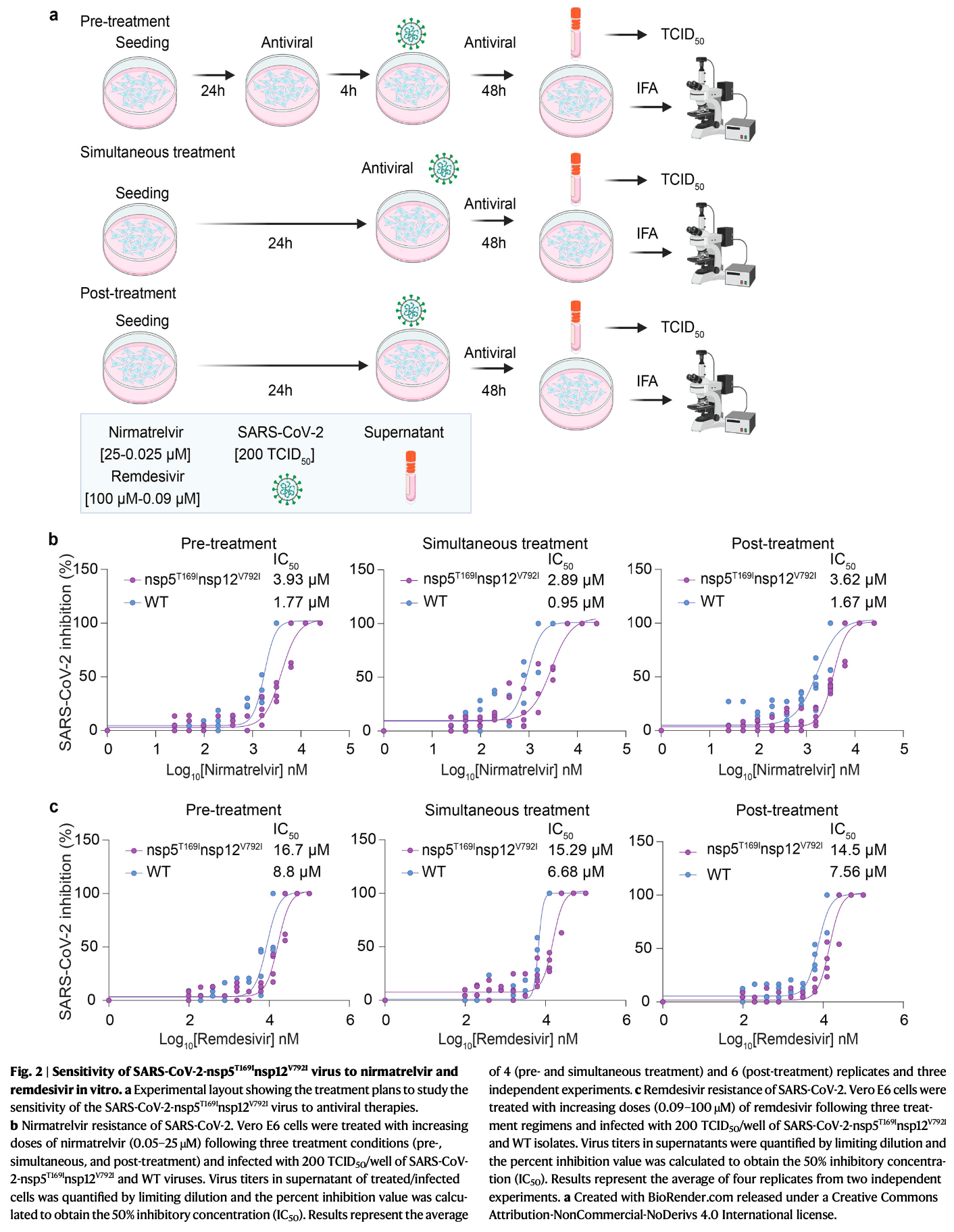
Analysis of 15 immunocompromised patients with persistent SARS-CoV-2 infection showing emergence of putative antiviral resistance mutations in nsp5 and nsp12 following treatment with remdesivir and nirmatrelvir-ritonavir. An infectious SARS-CoV-2 variant carrying nsp5 T169I and nsp12 V792I mutations was isolated from one patient more than 2 months after diagnosis, which transmitted efficiently in a hamster model. In vitro experiments confirmed this variant had decreased sensitivity to remdesivir and nirmatrelvir.
Resistance. Variants may be resistant to paxlovid1-8. Use may promote the emergence of variants that weaken host immunity and potentially contribute to long COVID9. Confounding by contraindication. Hoertel et al. find that over 50% of patients that died had a contraindication for the use of Paxlovid10. Retrospective studies that do not exclude contraindicated patients may significantly overestimate efficacy. Black box warning. The FDA notes that severe, life-threatening, and/or fatal adverse reactions due to drug interactions have been reported in patients treated with paxlovid11. Kidney and liver injury. Studies show significantly increased risk of acute kidney injury12 and liver injury13,14. Viral rebound. Studies show significantly increased risk of replication-competent viral rebound15-17.
Study covers remdesivir and paxlovid.
1.
Zhou et al., Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Science Advances, doi:10.1126/sciadv.add7197.
2.
Moghadasi et al., Rapid resistance profiling of SARS-CoV-2 protease inhibitors, npj Antimicrobials and Resistance, doi:10.1038/s44259-023-00009-0.
3.
Jochmans et al., The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir, mBio, doi:10.1128/mbio.02815-22.
4.
Lopez et al., SARS-CoV-2 Resistance to Small Molecule Inhibitors, Current Clinical Microbiology Reports, doi:10.1007/s40588-024-00229-6.
5.
Zvornicanin et al., Molecular Mechanisms of Drug Resistance and Compensation in SARS-CoV-2 Main Protease: The Interplay Between E166 and L50, bioRxiv, doi:10.1101/2025.01.24.634813.
6.
Vukovikj et al., Impact of SARS-CoV-2 variant mutations on susceptibility to monoclonal antibodies and antiviral drugs: a non-systematic review, April 2022 to October 2024, Eurosurveillance, doi:10.2807/1560-7917.ES.2025.30.10.2400252.
7.
Deschenes et al., Functional and structural characterization of treatment-emergent nirmatrelvir resistance mutations at low frequencies in the main protease (Mpro) reveals a unique evolutionary route for SARS-CoV-2 to gain resistance, The Journal of Infectious Diseases, doi:10.1093/infdis/jiaf294.
8.
Zhou (B) et al., SARS-CoV-2 Mpro inhibitor ensitrelvir: asymmetrical cross-resistance with nirmatrelvir and emerging resistance hotspots, Emerging Microbes & Infections, doi:10.1080/22221751.2025.2552716.
9.
Thomas et al., Nirmatrelvir-Resistant Mutations in SARS-CoV-2 Mpro Enhance Host Immune Evasion via Cleavage of NF-κB Essential Modulator, bioRxiv, doi:10.1101/2024.10.18.619137.
10.
Hoertel et al., Prevalence of Contraindications to Nirmatrelvir-Ritonavir Among Hospitalized Patients With COVID-19 at Risk for Progression to Severe Disease, JAMA Network Open, doi:10.1001/jamanetworkopen.2022.42140.
11.
FDA, Fact sheet for healthcare providers: emergency use authorization for paxlovid, www.fda.gov/media/155050/download.
12.
Kamo et al., Association of Antiviral Drugs for the Treatment of COVID-19 With Acute Renal Failure, In Vivo, doi:10.21873/invivo.13637.
13.
Wang et al., Development and validation of a nomogram to assess the occurrence of liver dysfunction in patients with COVID-19 pneumonia in the ICU, BMC Infectious Diseases, doi:10.1186/s12879-025-10684-1.
14.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
15.
Edelstein et al., SARS-CoV-2 virologic rebound with nirmatrelvir-ritonavir therapy, medRxiv, doi:10.1101/2023.06.23.23288598.
Nooruzzaman et al., 18 Sep 2024, retrospective, USA, peer-reviewed, 18 authors.
Contact: elodie.ghedin@nih.gov, dgdiel@cornell.edu, mis2053@med.cornell.edu.
Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections
Nature Communications, doi:10.1038/s41467-024-51924-3
We investigated the impact of antiviral treatment on the emergence of SARS-CoV-2 resistance during persistent infections in immunocompromised patients (n = 15). All patients received remdesivir and some also received nirmatrelvir-ritonavir (n = 3) or therapeutic monoclonal antibodies (n = 4). Sequence analysis showed that nine patients carried viruses with mutations in the nsp12 (RNA dependent RNA polymerase), while four had viruses with nsp5 (3C protease) mutations. Infectious SARS-CoV-2 with a double mutation in nsp5 (T169I) and nsp12 (V792I) was recovered from respiratory secretions 77 days after initial COVID-19 diagnosis from a patient sequentially treated with nirmatrelvir-ritonavir and remdesivir. In vitro characterization confirmed its decreased sensitivity to remdesivir and nirmatrelvir, which was overcome by combined antiviral treatment. Studies in golden Syrian hamsters demonstrated efficient transmission to contact animals. This study documents the isolation of SARS-CoV-2 carrying resistance mutations to both nirmatrelvir and remdesivir from a patient and demonstrates its transmissibility in vivo. Infection with SARS-CoV-2 in immunocompromised patients poses major clinical, therapeutic, and public health challenges. These patients often experience more severe infection outcome(s) than the general population with disease progression being influenced by the treatment of the underlying condition 1 . Moreover, while most people with a competent immune system successfully clear SARS-CoV-2 infection within days, immunocompromised patients may become persistently infected and present prolonged virus replication and shedding. Long-term viral replication contributes to intra-host evolution leading to the emergence of variants with mutations in the virus
Nucleic acid isolation and real-time reverse transcriptase PCR (rRT-PCR) Nucleic acid was extracted from oropharyngeal swabs and tissues collected at necropsy. A 10% (w/v) homogenate was prepared in DMEM from tissues (nasal turbinate, trachea, and lungs) using a stomacher (one speed cycle of 60 s, Stomacher® 80 Biomaster). The tissue homogenate was clarified by centrifuging at 2000×g for 10 min. In total, 200 µL of oropharyngeal swabs and clarified tissue homogenate was used for RNA extraction using the MagMax Core extraction kit (Thermo Fisher, Waltham, MA, USA) and the automated KingFisher Flex nucleic acid extractor (Thermo Fisher, Waltham, MA, USA). The rRT-PCR for total viral RNA detection was performed using the EZ-SARS-CoV-2 Real-Time RT-PCR assay (Tetracore Inc., Rockville, MD, USA), which detects both genomic and subgenomic viral RNA targeting the viral nucleoprotein gene. An internal inhibition control was included in all reactions. Positive and negative amplification controls were run side-by-side with test samples. Relative viral genome copy numbers were calculated based on the standard curve and determined using GraphPad Prism 9 (GraphPad, La Jolla, CA, USA). The amount of viral RNA detected in samples was expressed as log (genome copy number) per mL.
Virus isolation and titration All oropharyngeal swabs and tissue homogenates were subjected to virus isolation under Biosafety Level 3 (BSL-3) conditions at the Animal Health Diagnostic Center (ADHC) Research..
References
Aksamentov, Roemer, Hodcroft, Neher, Nextclade: clade assignment, mutation calling and quality control for viral genomes, J. Open Source Softw
Andrés, Emergence of Delta and Omicron variants carrying resistance-associated mutations in immunocompromised patients undergoing sotrovimab treatment with long-term viral excretion, Clin. Microbiol. Infect
Beigel, Remdesivir for the treatment of Covid-19-final report, New Engl. J. Med
Birnie, Development of resistance-associated mutations after sotrovimab administration in high-risk individuals infected with the SARS-CoV-2 Omicron variant, J. Am. Med. Assoc
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics
Chen, CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants, Bioinformatics
Dallakyan, Olson, Small-molecule library screening by docking with PyRx, Methods Mol. Biol
Duan, Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir, Nature
Gonzalez-Reiche, Introductions and early spread of SARS-CoV-2 in the New York City area, Science
Gottlieb, Early remdesivir to prevent progression to severe covid-19 in outpatients, New Engl. J. Med
Grubaugh, An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar, Genome Biol
Hedskog, Viral resistance analyses from the remdesivir phase 3 adaptive COVID-19 treatment trial-1 (ACTT-1), J. Infect. Dis
Heyer, Remdesivir-induced emergence of SARS-CoV2 variants in patients with prolonged infection, Cell Rep. Med
Hirotsu, Multidrug-resistant mutations to antiviral and antibody therapy in an immunocompromised patient infected with SARS-CoV-2, Med
Hu, Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, ACS Cent. Sci
Huygens, Oude Munnink, Gharbharan, Koopmans, Rijnders, Sotrovimab resistance and viral persistence after treatment of immunocompromised patients infected with the severe acute respiratory syndrome coronavirus 2 Omicron variant, Clin. Infect. Dis
Iketani, Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature
Ip, Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance, EBioMedicine
Kemp, SARS-CoV-2 evolution during treatment of chronic infection, Nature
Kim, PubChem 2023 update, Nucleic Acids Res
Kovalevsky, Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors, J. Biol. Chem
Kusakabe, Fungal microbiota sustains lasting immune activation of neutrophils and their progenitors in severe COVID-19, Nat. Immunol
Laskowski, Macarthur, Moss, Thornton, PROCHECK: a program to check the stereochemical quality of protein structures, J. Appl Crystallogr
Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM
Li, The Sequence Alignment/Map format and SAMtools, Bioinformatics
Marques, SARS-CoV-2 evolution during prolonged infection in immunocompromised patients, mBio
Martins, The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection, J. Virol
Martínez-López, COVID-19 severity and survival over time in patients with hematologic malignancies: a Population-Based Registry Study, Cancers
Mikulska, Triple combination therapy with 2 antivirals and monoclonal antibodies for persistent or relapsed severe acute respiratory syndrome coronavirus 2 infection in immunocompromised patients, Clin. Infect. Dis
Moghadasi, Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors, Sci. Adv
Najjar-Debbiny, Effectiveness of molnupiravir in high-risk patients: a propensity score matched analysis, Clin. Infect. Dis
Orth, Early combination therapy of COVID-19 in high-risk patients, Infection
Ou, Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events, Signal Transduct. Target Ther
Paraskevis, Real-world effectiveness of molnupiravir and nirmatrelvir/ritonavir as treatments for COVID-19 in patients at high risk, J. Infect. Dis
Pond, Martin, Anti-COVID drug accelerates viral evolution, Nature
Raglow, SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the omicron period: a multicentre, prospective analysis, Lancet Microbe
Rendeiro, Metabolic and immune markers for precise monitoring of COVID-19 severity and treatment, Front. Immunol
Rendeiro, Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression, Life Sci. Alliance
Rockett, Resistance mutations in SARS-CoV-2 delta variant after sotrovimab use, New Engl. J. Med
Roder, Optimized quantification of intra-host viral diversity in SARS-CoV-2 and influenza virus sequence data, mBio
Sanderson, A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature
Schrödinger, Delano, Pymol, None
Smithgall, Scherberkova, Whittier, Green, Comparison of cepheid Xpert Xpress and Abbott ID now to Roche Cobas for the rapid detection of SARS-CoV-2, J. Clin. Virol
Spinner, Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19, J. Am. Med. Assoc
Steffen, AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility, J. Comput. Chem
Stevens, Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms, Sci. Transl. Med
Tian, Efficacy and safety of paxlovid (nirmatrelvir/ritonavir) in the treatment of COVID-19: an updated meta-analysis and trial sequential analysis, Rev. Med. Virol
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem, doi:10.1002/jcc.21334
Van Der Auwera, O'connor, Genomics in The Cloud: Using Docker, GATK, and WDL in Terra
Wallace, Laskowski, Thornton, Ligplot: a program to generate schematic diagrams of protein-ligand interactions, Protein Eng. Des. Selection
Waterhouse, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res
Wilkinson, Recurrent SARS-CoV-2 mutations in immunodeficient patients, Virus Evol
Yang, Transient SARS-CoV-2 RNA-dependent RNA polymerase mutations after remdesivir treatment for chronic COVID-19 in two transplant recipients: case report and intra-host viral genomic investigation, Microorganisms
Yin, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science
Zhou, Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Sci. Adv
Zuckerman, Bucris, Keidar-Friedman, Amsalem, Brosh-Nissimov, Nirmatrelvir resistance-de novo E166V/L50V mutations in an immunocompromised patient treated with prolonged nirmatrelvir/ritonavir monotherapy leading to clinical and virological treatment failure-a case report, Clin. Infect. Dis
DOI record:
{
"DOI": "10.1038/s41467-024-51924-3",
"ISSN": [
"2041-1723"
],
"URL": "http://dx.doi.org/10.1038/s41467-024-51924-3",
"abstract": "<jats:title>Abstract</jats:title><jats:p>We investigated the impact of antiviral treatment on the emergence of SARS-CoV-2 resistance during persistent infections in immunocompromised patients (<jats:italic>n</jats:italic> = 15). All patients received remdesivir and some also received nirmatrelvir-ritonavir (<jats:italic>n</jats:italic> = 3) or therapeutic monoclonal antibodies (<jats:italic>n</jats:italic> = 4). Sequence analysis showed that nine patients carried viruses with mutations in the nsp12 (RNA dependent RNA polymerase), while four had viruses with nsp5 (3C protease) mutations. Infectious SARS-CoV-2 with a double mutation in nsp5 (T169I) and nsp12 (V792I) was recovered from respiratory secretions 77 days after initial COVID-19 diagnosis from a patient sequentially treated with nirmatrelvir-ritonavir and remdesivir. In vitro characterization confirmed its decreased sensitivity to remdesivir and nirmatrelvir, which was overcome by combined antiviral treatment. Studies in golden Syrian hamsters demonstrated efficient transmission to contact animals. This study documents the isolation of SARS-CoV-2 carrying resistance mutations to both nirmatrelvir and remdesivir from a patient and demonstrates its transmissibility in vivo.</jats:p>",
"alternative-id": [
"51924"
],
"article-number": "7999",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "13 June 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "21 August 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "18 September 2024"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Nooruzzaman",
"given": "Mohammed",
"sequence": "first"
},
{
"affiliation": [],
"family": "Johnson",
"given": "Katherine E. E.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rani",
"given": "Ruchi",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7363-2517",
"affiliation": [],
"authenticated-orcid": false,
"family": "Finkelsztein",
"given": "Eli J.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1643-8560",
"affiliation": [],
"authenticated-orcid": false,
"family": "Caserta",
"given": "Leonardo C.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6273-0592",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kodiyanplakkal",
"given": "Rosy P.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Wei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hsu",
"given": "Jingmei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Salpietro",
"given": "Maria T.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Banakis",
"given": "Stephanie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Albert",
"given": "Joshua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Westblade",
"given": "Lars F.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5043-8033",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zanettini",
"given": "Claudio",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7336-8071",
"affiliation": [],
"authenticated-orcid": false,
"family": "Marchionni",
"given": "Luigi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Soave",
"given": "Rosemary",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1515-725X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ghedin",
"given": "Elodie",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3237-8940",
"affiliation": [],
"authenticated-orcid": false,
"family": "Diel",
"given": "Diego G.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8296-0376",
"affiliation": [],
"authenticated-orcid": false,
"family": "Salvatore",
"given": "Mirella",
"sequence": "additional"
}
],
"container-title": "Nature Communications",
"container-title-short": "Nat Commun",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
9,
19
]
],
"date-time": "2024-09-19T21:05:46Z",
"timestamp": 1726779946000
},
"deposited": {
"date-parts": [
[
2024,
9,
19
]
],
"date-time": "2024-09-19T21:07:58Z",
"timestamp": 1726780078000
},
"indexed": {
"date-parts": [
[
2024,
9,
20
]
],
"date-time": "2024-09-20T04:31:31Z",
"timestamp": 1726806691582
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
9,
18
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
9,
18
]
],
"date-time": "2024-09-18T00:00:00Z",
"timestamp": 1726617600000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
9,
18
]
],
"date-time": "2024-09-18T00:00:00Z",
"timestamp": 1726617600000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41467-024-51924-3.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-024-51924-3",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-024-51924-3.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2024,
9,
18
]
]
},
"published-online": {
"date-parts": [
[
2024,
9,
18
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.3390/cancers15051497",
"author": "J Martínez-López",
"doi-asserted-by": "publisher",
"first-page": "1497",
"journal-title": "Cancers",
"key": "51924_CR1",
"unstructured": "Martínez-López, J. et al. COVID-19 severity and survival over time in patients with hematologic malignancies: a Population-Based Registry Study. Cancers 15, 1497 (2023).",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.1128/mbio.00110-24",
"author": "AD Marques",
"doi-asserted-by": "publisher",
"first-page": "e0011024",
"journal-title": "mBio",
"key": "51924_CR2",
"unstructured": "Marques, A. D. et al. SARS-CoV-2 evolution during prolonged infection in immunocompromised patients. mBio 15, e0011024 (2024).",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1002/rmv.2473",
"author": "H Tian",
"doi-asserted-by": "publisher",
"first-page": "e2473",
"journal-title": "Rev. Med. Virol.",
"key": "51924_CR3",
"unstructured": "Tian, H. et al. Efficacy and safety of paxlovid (nirmatrelvir/ritonavir) in the treatment of COVID‐19: an updated meta‐analysis and trial sequential analysis. Rev. Med. Virol. 33, e2473 (2023).",
"volume": "33",
"year": "2023"
},
{
"DOI": "10.1056/NEJMoa2116846",
"author": "RL Gottlieb",
"doi-asserted-by": "publisher",
"first-page": "305",
"journal-title": "New Engl. J. Med.",
"key": "51924_CR4",
"unstructured": "Gottlieb, R. L. et al. Early remdesivir to prevent progression to severe covid-19 in outpatients. New Engl. J. Med. 386, 305–315 (2022).",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1056/NEJMoa2007764",
"author": "JH Beigel",
"doi-asserted-by": "publisher",
"first-page": "1813",
"journal-title": "New Engl. J. Med.",
"key": "51924_CR5",
"unstructured": "Beigel, J. H. et al. Remdesivir for the treatment of Covid-19—final report. New Engl. J. Med. 383, 1813–1826 (2020).",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1001/jama.2020.16349",
"author": "CD Spinner",
"doi-asserted-by": "publisher",
"first-page": "1048",
"journal-title": "J. Am. Med. Assoc.",
"key": "51924_CR6",
"unstructured": "Spinner, C. D. et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19. J. Am. Med. Assoc. 324, 1048 (2020).",
"volume": "324",
"year": "2020"
},
{
"DOI": "10.1093/infdis/jiad324",
"author": "D Paraskevis",
"doi-asserted-by": "publisher",
"first-page": "1667",
"journal-title": "J. Infect. Dis.",
"key": "51924_CR7",
"unstructured": "Paraskevis, D. et al. Real-world effectiveness of molnupiravir and nirmatrelvir/ritonavir as treatments for COVID-19 in patients at high risk. J. Infect. Dis. 228, 1667–1674 (2023).",
"volume": "228",
"year": "2023"
},
{
"DOI": "10.1093/cid/ciac781",
"author": "R Najjar-Debbiny",
"doi-asserted-by": "publisher",
"first-page": "453",
"journal-title": "Clin. Infect. Dis.",
"key": "51924_CR8",
"unstructured": "Najjar-Debbiny, R. et al. Effectiveness of molnupiravir in high-risk patients: a propensity score matched analysis. Clin. Infect. Dis. 76, 453–460 (2023).",
"volume": "76",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06649-6",
"author": "T Sanderson",
"doi-asserted-by": "publisher",
"first-page": "594",
"journal-title": "Nature",
"key": "51924_CR9",
"unstructured": "Sanderson, T. et al. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature 623, 594–600 (2023).",
"volume": "623",
"year": "2023"
},
{
"DOI": "10.1016/j.xcrm.2022.100735",
"author": "A Heyer",
"doi-asserted-by": "publisher",
"first-page": "100735",
"journal-title": "Cell Rep. Med.",
"key": "51924_CR10",
"unstructured": "Heyer, A. et al. Remdesivir-induced emergence of SARS-CoV2 variants in patients with prolonged infection. Cell Rep. Med. 3, 100735 (2022).",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1038/d41586-023-03248-3",
"author": "SL Kosakovsky Pond",
"doi-asserted-by": "publisher",
"first-page": "486",
"journal-title": "Nature",
"key": "51924_CR11",
"unstructured": "Kosakovsky Pond, S. L. & Martin, D. Anti-COVID drug accelerates viral evolution. Nature 623, 486–487 (2023).",
"volume": "623",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06609-0",
"author": "Y Duan",
"doi-asserted-by": "publisher",
"first-page": "376",
"journal-title": "Nature",
"key": "51924_CR12",
"unstructured": "Duan, Y. et al. Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir. Nature 622, 376–382 (2023).",
"volume": "622",
"year": "2023"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"author": "S Iketani",
"doi-asserted-by": "publisher",
"first-page": "558",
"journal-title": "Nature",
"key": "51924_CR13",
"unstructured": "Iketani, S. et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 613, 558–564 (2023).",
"volume": "613",
"year": "2023"
},
{
"DOI": "10.3390/microorganisms11082096",
"author": "S Yang",
"doi-asserted-by": "publisher",
"first-page": "2096",
"journal-title": "Microorganisms",
"key": "51924_CR14",
"unstructured": "Yang, S. et al. Transient SARS-CoV-2 RNA-dependent RNA polymerase mutations after remdesivir treatment for chronic COVID-19 in two transplant recipients: case report and intra-host viral genomic investigation. Microorganisms 11, 2096 (2023).",
"volume": "11",
"year": "2023"
},
{
"DOI": "10.1126/scitranslmed.abo0718",
"author": "LJ Stevens",
"doi-asserted-by": "publisher",
"first-page": "eabo0718",
"journal-title": "Sci. Transl. Med",
"key": "51924_CR15",
"unstructured": "Stevens, L. J. et al. Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 14, eabo0718 (2022).",
"volume": "14",
"year": "2022"
},
{
"key": "51924_CR16",
"unstructured": "https://www.gilead.com/-/media/files/pdfs/medicines/covid-19/veklury/veklury_pi.pdf (2024)."
},
{
"DOI": "10.1016/S2666-5247(23)00336-1",
"author": "Z Raglow",
"doi-asserted-by": "publisher",
"first-page": "e235",
"journal-title": "Lancet Microbe",
"key": "51924_CR17",
"unstructured": "Raglow, Z. et al. SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the omicron period: a multicentre, prospective analysis. Lancet Microbe 5, e235–e246 (2024).",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1038/s41586-021-03291-y",
"author": "SA Kemp",
"doi-asserted-by": "publisher",
"first-page": "277",
"journal-title": "Nature",
"key": "51924_CR18",
"unstructured": "Kemp, S. A. et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592, 277–282 (2021).",
"volume": "592",
"year": "2021"
},
{
"DOI": "10.1038/s41392-022-00992-2",
"author": "J Ou",
"doi-asserted-by": "publisher",
"first-page": "138",
"journal-title": "Signal Transduct. Target Ther.",
"key": "51924_CR19",
"unstructured": "Ou, J. et al. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Target Ther. 7, 138 (2022).",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1093/cid/ciac601",
"doi-asserted-by": "crossref",
"key": "51924_CR20",
"unstructured": "Huygens, S., Oude Munnink, B., Gharbharan, A., Koopmans, M. & Rijnders, B. Sotrovimab resistance and viral persistence after treatment of immunocompromised patients infected with the severe acute respiratory syndrome coronavirus 2 Omicron variant. Clin. Infect. Dis. 76, e507–e509 (2023)."
},
{
"DOI": "10.1016/j.cmi.2022.08.021",
"author": "C Andrés",
"doi-asserted-by": "publisher",
"first-page": "240",
"journal-title": "Clin. Microbiol. Infect.",
"key": "51924_CR21",
"unstructured": "Andrés, C. et al. Emergence of Delta and Omicron variants carrying resistance-associated mutations in immunocompromised patients undergoing sotrovimab treatment with long-term viral excretion. Clin. Microbiol. Infect. 29, 240–246 (2023).",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.1001/jama.2022.13854",
"author": "E Birnie",
"doi-asserted-by": "publisher",
"first-page": "1104",
"journal-title": "J. Am. Med. Assoc.",
"key": "51924_CR22",
"unstructured": "Birnie, E. et al. Development of resistance-associated mutations after sotrovimab administration in high-risk individuals infected with the SARS-CoV-2 Omicron variant. J. Am. Med. Assoc. 328, 1104 (2022).",
"volume": "328",
"year": "2022"
},
{
"DOI": "10.1056/NEJMc2120219",
"author": "R Rockett",
"doi-asserted-by": "publisher",
"first-page": "1477",
"journal-title": "New Engl. J. Med.",
"key": "51924_CR23",
"unstructured": "Rockett, R. et al. Resistance mutations in SARS-CoV-2 delta variant after sotrovimab use. New Engl. J. Med. 386, 1477–1479 (2022).",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1093/infdis/jiad270",
"author": "C Hedskog",
"doi-asserted-by": "publisher",
"first-page": "1263",
"journal-title": "J. Infect. Dis.",
"key": "51924_CR24",
"unstructured": "Hedskog, C. et al. Viral resistance analyses from the remdesivir phase 3 adaptive COVID-19 treatment trial-1 (ACTT-1). J. Infect. Dis. 228, 1263–1273 (2023).",
"volume": "228",
"year": "2023"
},
{
"DOI": "10.1093/ve/veac050",
"author": "SAJ Wilkinson",
"doi-asserted-by": "publisher",
"first-page": "veac050",
"journal-title": "Virus Evol.",
"key": "51924_CR25",
"unstructured": "Wilkinson, S. A. J. et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 8, veac050 (2022).",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1021/acscentsci.3c00538",
"author": "Y Hu",
"doi-asserted-by": "publisher",
"first-page": "1658",
"journal-title": "ACS Cent. Sci.",
"key": "51924_CR26",
"unstructured": "Hu, Y. et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. ACS Cent. Sci. 9, 1658–1669 (2023).",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1016/j.ebiom.2023.104559",
"author": "JD Ip",
"doi-asserted-by": "publisher",
"first-page": "104559",
"journal-title": "EBioMedicine",
"key": "51924_CR27",
"unstructured": "Ip, J. D. et al. Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance. EBioMedicine 91, 104559 (2023).",
"volume": "91",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.add7197",
"author": "Y Zhou",
"doi-asserted-by": "publisher",
"first-page": "eadd7197",
"journal-title": "Sci. Adv.",
"key": "51924_CR28",
"unstructured": "Zhou, Y. et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system. Sci. Adv. 8, eadd7197 (2022).",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1093/cid/ciad494",
"author": "NS Zuckerman",
"doi-asserted-by": "publisher",
"first-page": "352",
"journal-title": "Clin. Infect. Dis.",
"key": "51924_CR29",
"unstructured": "Zuckerman, N. S., Bucris, E., Keidar-Friedman, D., Amsalem, M. & Brosh-Nissimov, T. Nirmatrelvir resistance—de novo E166V/L50V mutations in an immunocompromised patient treated with prolonged nirmatrelvir/ritonavir monotherapy leading to clinical and virological treatment failure—a case report. Clin. Infect. Dis. 78, 352–355 (2024).",
"volume": "78",
"year": "2024"
},
{
"DOI": "10.1126/sciadv.ade8778",
"author": "SA Moghadasi",
"doi-asserted-by": "publisher",
"first-page": "eade8778",
"journal-title": "Sci. Adv.",
"key": "51924_CR30",
"unstructured": "Moghadasi, S. A. et al. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. Sci. Adv. 9, eade8778 (2023).",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1007/s15010-023-02125-5",
"author": "HM Orth",
"doi-asserted-by": "publisher",
"first-page": "877",
"journal-title": "Infection",
"key": "51924_CR31",
"unstructured": "Orth, H. M. et al. Early combination therapy of COVID-19 in high-risk patients. Infection 52, 877–889 (2024).",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1093/cid/ciad181",
"author": "M Mikulska",
"doi-asserted-by": "publisher",
"first-page": "280",
"journal-title": "Clin. Infect. Dis.",
"key": "51924_CR32",
"unstructured": "Mikulska, M. et al. Triple combination therapy with 2 antivirals and monoclonal antibodies for persistent or relapsed severe acute respiratory syndrome coronavirus 2 infection in immunocompromised patients. Clin. Infect. Dis. 77, 280–286 (2023).",
"volume": "77",
"year": "2023"
},
{
"DOI": "10.1016/j.medj.2023.08.001",
"author": "Y Hirotsu",
"doi-asserted-by": "publisher",
"first-page": "813",
"journal-title": "Med",
"key": "51924_CR33",
"unstructured": "Hirotsu, Y. et al. Multidrug-resistant mutations to antiviral and antibody therapy in an immunocompromised patient infected with SARS-CoV-2. Med. 4, 813–824.e4 (2023).",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2021.809937",
"author": "AF Rendeiro",
"doi-asserted-by": "publisher",
"first-page": "809937",
"journal-title": "Front. Immunol.",
"key": "51924_CR34",
"unstructured": "Rendeiro, A. F. et al. Metabolic and immune markers for precise monitoring of COVID-19 severity and treatment. Front. Immunol. 12, 809937 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.26508/lsa.202000955",
"author": "AF Rendeiro",
"doi-asserted-by": "publisher",
"first-page": "e202000955",
"journal-title": "Life Sci. Alliance",
"key": "51924_CR35",
"unstructured": "Rendeiro, A. F. et al. Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression. Life Sci. Alliance 4, e202000955 (2021).",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.1038/s41590-023-01637-4",
"author": "T Kusakabe",
"doi-asserted-by": "publisher",
"first-page": "1879",
"journal-title": "Nat. Immunol.",
"key": "51924_CR36",
"unstructured": "Kusakabe, T. et al. Fungal microbiota sustains lasting immune activation of neutrophils and their progenitors in severe COVID-19. Nat. Immunol. 24, 1879–1889 (2023).",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/j.jcv.2020.104428",
"author": "MC Smithgall",
"doi-asserted-by": "publisher",
"first-page": "104428",
"journal-title": "J. Clin. Virol.",
"key": "51924_CR37",
"unstructured": "Smithgall, M. C., Scherberkova, I., Whittier, S. & Green, D. A. Comparison of cepheid Xpert Xpress and Abbott ID now to Roche Cobas for the rapid detection of SARS-CoV-2. J. Clin. Virol. 128, 104428 (2020).",
"volume": "128",
"year": "2020"
},
{
"DOI": "10.1126/science.abc1917",
"author": "AS Gonzalez-Reiche",
"doi-asserted-by": "publisher",
"first-page": "297",
"journal-title": "Science",
"key": "51924_CR38",
"unstructured": "Gonzalez-Reiche, A. S. et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science 369, 297–301 (2020).",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1128/mbio.01046-23",
"author": "AE Roder",
"doi-asserted-by": "publisher",
"first-page": "e0104623",
"journal-title": "mBio",
"key": "51924_CR39",
"unstructured": "Roder, A. E. et al. Optimized quantification of intra-host viral diversity in SARS-CoV-2 and influenza virus sequence data. mBio 14, e0104623 (2023).",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1093/bioinformatics/btp352",
"author": "H Li",
"doi-asserted-by": "publisher",
"first-page": "2078",
"journal-title": "Bioinformatics",
"key": "51924_CR40",
"unstructured": "Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).",
"volume": "25",
"year": "2009"
},
{
"key": "51924_CR41",
"unstructured": "Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013)."
},
{
"DOI": "10.1093/bioinformatics/btu170",
"author": "AM Bolger",
"doi-asserted-by": "publisher",
"first-page": "2114",
"journal-title": "Bioinformatics",
"key": "51924_CR42",
"unstructured": "Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1186/s13059-018-1618-7",
"author": "ND Grubaugh",
"doi-asserted-by": "publisher",
"journal-title": "Genome Biol.",
"key": "51924_CR43",
"unstructured": "Grubaugh, N. D. et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 20, 8 (2019).",
"volume": "20",
"year": "2019"
},
{
"key": "51924_CR44",
"unstructured": "Van der Auwera, G. A. & O’Connor, B. D. Genomics in The Cloud: Using Docker, GATK, and WDL in Terra (O’Reilly Media, 2020)."
},
{
"DOI": "10.21105/joss.03773",
"author": "I Aksamentov",
"doi-asserted-by": "publisher",
"first-page": "3773",
"journal-title": "J. Open Source Softw.",
"key": "51924_CR45",
"unstructured": "Aksamentov, I., Roemer, C., Hodcroft, E. & Neher, R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 6, 3773 (2021).",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1093/bioinformatics/btab856",
"author": "C Chen",
"doi-asserted-by": "publisher",
"first-page": "1735",
"journal-title": "Bioinformatics",
"key": "51924_CR46",
"unstructured": "Chen, C. et al. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 38, 1735–1737 (2022).",
"volume": "38",
"year": "2022"
},
{
"DOI": "10.1128/jvi.00961-22",
"author": "M Martins",
"doi-asserted-by": "publisher",
"first-page": "e0096122",
"journal-title": "J. Virol.",
"key": "51924_CR47",
"unstructured": "Martins, M. et al. The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection. J. Virol. 96, e0096122 (2022).",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/j.jbc.2023.104886",
"author": "A Kovalevsky",
"doi-asserted-by": "publisher",
"first-page": "104886",
"journal-title": "J. Biol. Chem.",
"key": "51924_CR48",
"unstructured": "Kovalevsky, A. et al. Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors. J. Biol. Chem. 299, 104886 (2023).",
"volume": "299",
"year": "2023"
},
{
"DOI": "10.1126/science.abc1560",
"author": "W Yin",
"doi-asserted-by": "publisher",
"first-page": "1499",
"journal-title": "Science",
"key": "51924_CR49",
"unstructured": "Yin, W. et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 (2020).",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1093/nar/gky427",
"author": "A Waterhouse",
"doi-asserted-by": "publisher",
"first-page": "W296",
"journal-title": "Nucleic Acids Res.",
"key": "51924_CR50",
"unstructured": "Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.1107/S0021889892009944",
"author": "RA Laskowski",
"doi-asserted-by": "publisher",
"first-page": "283",
"journal-title": "J. Appl Crystallogr",
"key": "51924_CR51",
"unstructured": "Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl Crystallogr 26, 283–291 (1993).",
"volume": "26",
"year": "1993"
},
{
"key": "51924_CR52",
"unstructured": "Schrödinger, L., & DeLano, W. PyMOL. Retrieved from http://www.pymol.org/pymol (2020)."
},
{
"DOI": "10.1002/jcc.21256",
"doi-asserted-by": "crossref",
"key": "51924_CR53",
"unstructured": "Steffen, C. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 31, 2785–2791 (2010)."
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "publisher",
"key": "51924_CR54",
"unstructured": "Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. https://doi.org/10.1002/jcc.21334 (2009)."
},
{
"DOI": "10.1093/nar/gkac956",
"author": "S Kim",
"doi-asserted-by": "publisher",
"first-page": "D1373",
"journal-title": "Nucleic Acids Res.",
"key": "51924_CR55",
"unstructured": "Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–D1380 (2023).",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1007/978-1-4939-2269-7_19",
"author": "S Dallakyan",
"doi-asserted-by": "publisher",
"first-page": "243",
"journal-title": "Methods Mol. Biol.",
"key": "51924_CR56",
"unstructured": "Dallakyan, S. & Olson, A. J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263, 243–250 (2015).",
"volume": "1263",
"year": "2015"
},
{
"DOI": "10.1093/protein/8.2.127",
"author": "AC Wallace",
"doi-asserted-by": "publisher",
"first-page": "127",
"journal-title": "Protein Eng. Des. Selection",
"key": "51924_CR57",
"unstructured": "Wallace, A. C., Laskowski, R. A. & Thornton, J. M. Ligplot: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Selection 8, 127–134 (1995).",
"volume": "8",
"year": "1995"
}
],
"reference-count": 57,
"references-count": 57,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41467-024-51924-3"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "15"
}
nooruzzaman