Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study
et al., Research Square, doi:10.21203/rs.3.rs-628277/v1, Jun 2021
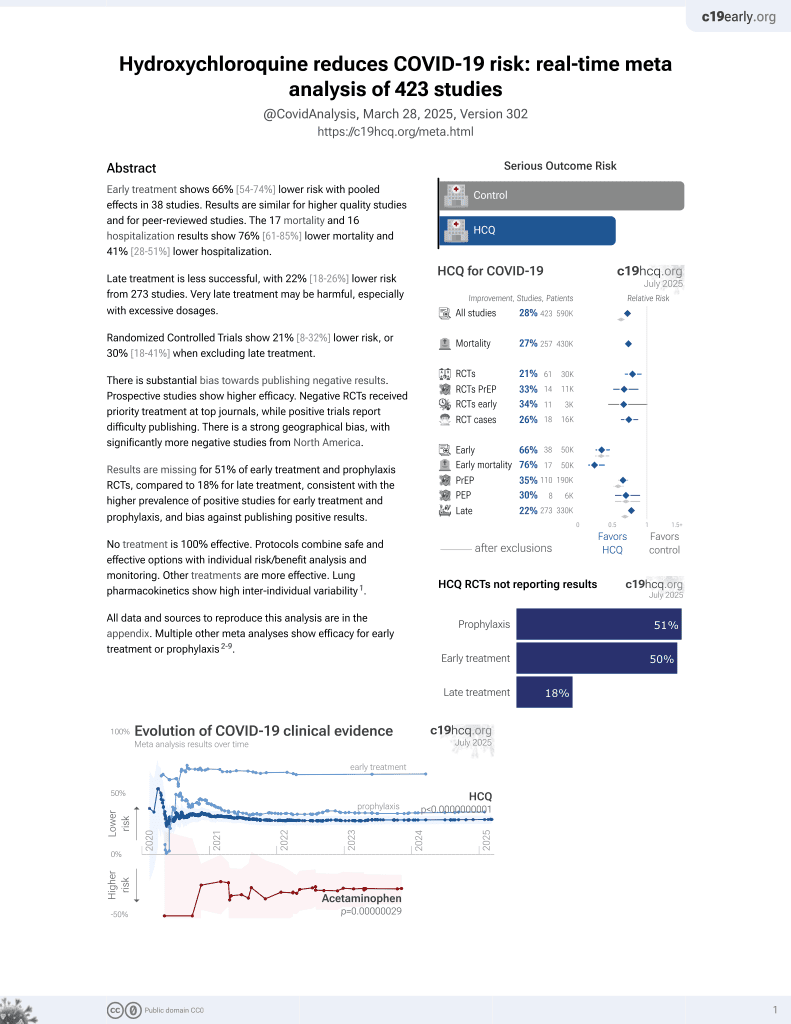
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 424 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico study showing stronger inhibition of SAR-CoV-2 for HCQ+favipiravir+oseltamivir compared to any of these alone or combinations of two of these drugs.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Yadav et al., 21 Jun 2021, preprint, 2 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study
doi:10.21203/rs.3.rs-628277/v1
The virus SARS-CoV-2 has created a situation of global emergency all over the world from the last few months. We are witnessing a helpless situation due to COVID-19 as no vaccine or drug is effective against the disease. In the present study, we have tested the repurposing efficacy of some currently used combination drugs against COVID-19. We have tried to understand the mechanism of action of some repurposed drugs:Favipiravir (F), Hydroxychloroquine (H) and Oseltamivir (O). The ADME analysis have suggested strong inhibitory possibility of F, H, O combination towards receptor protein of 3CL pro of SARS-CoV-2 virus. The strong binding affinity, number of hydrogen bond interaction between inhibitor, receptor and lower inhibition constant computed from molecular docking validated the better complexation possibility of F+H+O:3CL pro combination. Various thermodynamical output from Molecular dynamics (MD) simulations like potential energy (Eg), temperature (T), density, pressure, SASA energy, interaction energies, Gibbs free energy (ΔGbind) etc., also favored the complexation between F+H+O and CoV-2 protease. Our in-silico results have recommended the strong candidature of combination drugs Favipiravir, Hydroxychloroquine and Oseltamivir as a potential lead inhibitor for targeting SARS-CoV-2 infections.
Supplementary Files This is a list of supplementary les associated with this preprint. Click to download.
ResearchHighlights.docx Supportingdocument.docx graphicalabstract.pdf
References
Anuj, Gaurav, Sanjeev, Mansi, Pankaj et al., Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1772112
Apparsundaram, Ferguson, Jr, Blakely, Molecular cloning of a human, hemicholinium-3-sensitive choline transporter, Biochem. Biophys. Res. Commun
Baron, Fons, Albrecht, Viral Pathogenesis
Becke, A new inhomogeneity parameter in density-functional theory, Journal of Chemical Physics, doi:10.1063/1.475007
Beigel, Remdesivir for the treatment of COVID-19-preliminary report, N Engl J Med, doi:10.1056/NEJMoa2007764
Ben-Zvi, Kivity, Langevitz, Hydroxychloroquine: From Malaria to Autoimmunity, Clinic Rev Allerg Immunol, doi:10.1007/s12016-010-8243-x
Benson, Daggett, A comparison of multiscale methods for the analysis of molecular dynamics simulations, The Journal of Physical Chemistry. B, doi:10.1021/jp302103t22494262
Berendsen, Van Der Spoel, Van Drunen, GROMACS: A message passing parallel molecular dynamics implementation, Computer Physics Communications, doi:10.1016/0010-4655(95)00042-E
Burley, Berman, Bhikadiya, Bi, Chen et al., RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy, Nucleic Acids Research, doi:10.1093/nar/gky1004
Cao, Deng, Dai, Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel medicine and infectious disease, doi:10.1016/j.tmaid.2020.101647
Chan, Yao, Yeung, Deng, Bao et al., Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset, The Journal of infectious diseases
Chang, Lo, Wang, Hou, Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein, Drug Discovery Today
Choudhary, Shaikh, Tul-Wahab, Ur-Rahman, In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation, Plos one
Chowdhury, Pathak, Neuroprotective Immunity by Essential Nutrient "Choline" for the Prevention of SARS CoV2 Infections: An In Silico Study by Molecular Dynamics Approach, Chemical Physics Letters, doi:10.1016/j.cplett.2020.138057
Chowdhuryp, In silico investigation of phytoconstituents from indian medicinal herb 'Tinospora Cordifolia (Giloy)' against SARS-CoV-2 (COVID-19) by molecular dynamics approach, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1803968
Costanzo, Giglio, SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus, Current Medicinal Chemistry, doi:10.2174/0929867327666200416131117
D'acquarica, Chiral switches of chloroquine and hydroxychloroquine: potential drugs to treat COVID-19, Drug Discovery Today, doi:10.1016/j.drudis.2020.04.021
Daina, Michielin, Zoete, Swiss ADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Sci. Rep, doi:10.1038/srep42717
Dennington, Keith, Millam, Gauss View, Version 4.1
Dey, Saini, Dhembla, Bhatt, Rajesh et al., Suramin, Penciclovir and Anidulafungin bind nsp12, which governs the RNA-dependent-RNA polymerase activity of SARS-CoV-2, with higher interaction energy than Remdesivir, indicating potential in the treatment of Covid-19 infection
Dharmendra, Molecular insights into the interaction of RONS and Thieno [3,2-c]pyran analogs with SIRT6/COX-2: a molecular dynamics study, Scientific Reports, doi:10.1038/s41598-018-22972-9
Doi, Ikeda, Hayase, Nafamostat mesylate treatment in combination with favipiravir for patients critically ill with Covid-19: a case series, Crit Care, doi:10.1186/s13054-020-03078-z
Dong, Li, Teng, Yin, Cao, Syntheses and neuroprotective effects evolution of oxymatrine derivatives as anti-Alzheimer's disease agents, Chemical Biology & Drug Design
Dyall, Coleman, Hart, Venkataramant, Holbrook et al., Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection, Antimicrobial
Elfiky, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARSCoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study, Life Sciences, doi:10.1016/j.lfs.2020.117592
Falzarano, Wit, Rasmussen, Feldmann, Okumura et al., Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques, Nature medicine
Frecer, Miertus, Antiviral agents against COVID-19: structure-based design of specific peptidomimetic inhibitors of SARS-CoV-2 main protease, RSC Advances
Fried, Shiffman, Reddy, Smith, Marinos et al., Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection, New England Journal of Medicine
Frisch, Gaussian 09
Furukawa, Brooks, Sobel, Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic, Emerg. Infect. Dis, doi:10.3201/eid2607.201595
Furuta, Takahashi, Fukuda, Kuno, Kamiyama et al., In vitro and in vivo activities of anti-influenza virus compound T-705, Antimicrob. Agents Chemother
Gautret, Lagier, Parola, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int J Antimicrob Agents, doi:10.1016/j.ijantimicag.2020.105949
Ghinai, First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA, The Lancet, doi:10.1016/S0140-6736(20)30607-3
Gorden, Gwendolynmj, Bouhaddou, A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing, doi:10.1101/2020.03.22.002386
Grant, Doxycycline prophylaxis for bacterial sexually transmitted infections, Clinical Infectious Diseases
Grein, Ohmagari, Shin, Diaz, Asperges et al., Compassionate use of remdesivir for patients with severe Covid-19, N. Engl. J. Med, doi:10.1056/NEJMoa2007016
Guan, Ni, -Y, Hu, Liang et al., Clinical characteristics of Coronavirus disease 2019 in China, The New England Journal of Medicine, doi:10.1056/NEJMoa2002032
Gunsteren, Billeter, Eising, Hünenberger, Krüger et al., Biomolecular simulation: The GROMOS96 manual and user guide
Gupta, Singh, Kushwaha, Prajapati, Shuaib et al., Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1776157
Harrison, Coronavirus puts drug repurposing on the fast track, Nature biotechnology
Hart, Dyall, Postnikova, Zhou, Kindrachuk et al., Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays, The Journal of general virology, doi:10.1099/vir.0.061911-0
He, Xiao, Yu, Song, Zhang et al., 2-(3-Hydroxybenzyl) benzo [d] isothiazol-3 (2H)-one Mannich base derivatives as potential multifunctional anti-Alzheimer's agents, Medicinal Chemistry Research
Heidary, Gharebaghi, Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen, The Journal of Antibiotics
Hoffmann, Schroeders, Kleine-Weber, Müller, Drosten et al., Nafamostat mesylate blocks activation of SARS-CoV-2: New treatment option for COVID-19, Antimicrobial Agents and Chemotherapy
Huang, Wang, Li, Ren, Zhao et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet
Hung, Lung, Tso, Liur, Chung et al., Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label,randomised, phase 2 trial, The Lancet
Hurt, Holien, Parker, Oseltamivir Resistance and the H274Y Neuraminidase Mutation in Seasonal. Pandemic and Highly Pathogenic Influenza Viruses, Drugs, doi:10.2165/11531450-000000000-00000
Jiang, Deng, Zhang, Cai, Cheungcw et al., Review of the clinical characteristics of coronavirus disease 2019 (COVID-19), Journal of General Internal Medicine, doi:10.1007/s11606-020-05762-w
Jin, Du, Xu, Deng, Liu et al., Structure of M pro from COVID-19 virus and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kapoor, Saigal, Elongavan, Action and resistance mechanisms of antibiotics: A guide for clinicians, J Anaesthesiol Clin Pharmacol, doi:10.4103/joacp.JOACP_349_15
Kaur, Charan, Dutta, Sharma, Bhardwaj et al., Favipiravir Use in COVID-19: Analysis of Suspected Adverse Drug Events Reported in the WHO Database. Infection and drug resistance
Keil, Ali, The avian flu: some lessons learned from the 2003 SARS outbreak in Toronto, Area
Kelleni, Nitazoxanide/Azithromycin combination for COVID-19: A suggested new protocol for COVID-19 early management
Kollman, Massova, Reyes, Kuhn, Huo et al., Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models, Acc. Chem. Res, doi:10.1021/ar000033j
Li, Asymptomatic and human-to-human transmission of SARS-CoV-2 in a 2-family cluster. Xuzhou, China, Emerg. Infect. Dis, doi:10.3201/eid2607.200718
Lipinski, Lead-and drug-like compounds: The rule-of-five revolution, Drug Discovery Today. Technologies, doi:10.1016/j.ddtec.2004.11.007
Martorana, Gentile, Lauria, In Silico Insights into the SARS CoV-2 Main Protease Suggest NADH Endogenous Defences in the Control of the Pandemic Coronavirus Infection, Viruses
Mcdowell, Nada, Sponer, Walter, Molecular Dynamics Simulations of RNA: An In Silico Single Molecule Approach, Biopolymers, doi:10.1002/bip.20620
Micco, Musella, Scala, Sala, Campiglia et al., In silico Analysis Revealed Potential Anti-SARS-CoV-2 Main Protease Activity by the Zonulin Inhibitor Larazotide Acetate, Frontiers in Chemistry
Morris, Huey, Lindstrom, Sannermf, Belew et al., AutoDock4 and Auto Dock Tools4: Automated docking with selective receptor flexibility, Journal of Computational Chemistry, doi:10.1002/jcc.21256
Muradrasoli, Balint, Wahlgren, Waldenstrom, Belak et al., Prevalence and Phylogeny of Coronaviruses in Wild Birds from the Bering Strait Area (Beringia), PLoS ONE, doi:10.1371/journal.pone.0013640
Ortega, Serrano, Pujolfh, Rangel, Role of changes in SARS-COV-2 spike protein in the interaction with the human ACE2 Receptor: an in silico analysis, EXCLI Journal, doi:10.17179/excli2020-1167
Pooja, Chowdhury, Influence of size and shape on optical and electronic properties of CdTe quantum dots in aqueous environment
Priyadarsini, Pandas, Singh, Beheraa, Biswal et al., In silico structural delineation of nucleocapsid protein of SARS-CoV-2, Journal of Entomology and Zoology Studies
Rainsford, Parke, Clifford-Rashotte, Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases, Inflammopharmacol, doi:10.1007/s10787-015-0239-y
Rana, Chowdhury, Perturbation of hydrogen bonding in hydrated pyrrole-2-carboxaldehyde complexes, Journal of molecular modeling
Rashmi, Rajendra, Andrew, g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations, J. Chem. Inf. Model, doi:10.1021/ci500020m
Rota, Oberste, Monroe, Nix, Campagnoli et al., Characterization of a novel coronavirus associated with severe acute respiratory syndrome, Science, doi:10.1126/science.1085952
Rothe, Schunk, Sothmann, Bretzel, Froeschl et al., Transmission of 2019-nCoV infection from an asymptomatic contact in Germany, New England, Journal of Medicine, doi:10.1056/NEJMc2001468
Selvaraj, Panwar, Dinesh, Boura, Singh et al., Microsecond MD Simulation and Multiple-Conformation Virtual Screening to Identify Potential Anti-COVID-19 Inhibitors Against SARS-CoV-2 Main Protease, Front. Chem, doi:10.3389/fchem
Sepaynayim, Anti-COVID-19 terpenoid from marine sources: A docking, admet and molecular dynamics study, Journal of molecular structure, doi:10.1016/j.molstruc.2020.129433
Shalhoub, Farahat, Simhairi, Shamma, Siddiqi et al., IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study, Journal of Antimicrobial Chemotherapy
Shannon, Selisko, Le, Huchting, Touret et al., Favipiravir strikes the SARS-CoV-2 at its Achilles heel. the RNA polymerase
Singh, Parida, Lingaraju, Kesavan, Kumar et al., Drug repurposing approach to fght COVID-19, Pharmacological Reports, doi:10.1007/s43440-020-00155-6
Stephen, DREIDING: a generic force field for molecular simulations, J. Phys. Chem, doi:10.1021/j100389a010
Su, Wong, Shi, Liu, Lai et al., Epidemiology, genetic recombination, and pathogenesis of coronaviruses, Trends Microbiol, doi:10.1016/j.tim.2016.03.003
Suryapad, Chi, Lees, Dietary therapy and herbal medicine for COVID-19 prevention: a review and perspective, J. Tradit. Complement. Med
Ter Meulen, Van Den, Brink, Poon, Marissen et al., Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants, PLoS Med
Thorlund, Dron, Park, Hsu, Forrest et al., A real-time dashboard of clinical trials for COVID-19, The Lancet Digital Health
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem, doi:10.1002/jcc.21334
Veber, Johnson, Cheng, Smith, Ward et al., Molecular properties that influence the oral bioavailability of drug candidates, J. Med. Chem
Velavan, Meyercg, The COVID-19 epidemic. Tropical Medicine & International Health, TM & IH, doi:10.1111/tmi.13383
Wang, Hu, Hu, Zhu, Liu et al., Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China, JAMA, doi:10.1001/jama.2020.1585
Woo, Lau, Chu, Chan, Tsoi et al., Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia, Journal of Virology, doi:10.1128/JVI.79.2.884-895.2005
Wu, Zhao, Yu, Chen, Song et al., A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Yang, Use of herbal drugs to treat COVID-19 should be with caution, The Lancet
Yin, Mao, Luan, Shen, Shen et al., Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by Remdesivir, Science
Zhou, Huang, Synthesis, anti-malaria and anti-bacterial activities of marine alkaloids, Chemical Biology & Drug Design
DOI record:
{
"DOI": "10.21203/rs.3.rs-628277/v1",
"URL": "http://dx.doi.org/10.21203/rs.3.rs-628277/v1",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The virus SARS-CoV-2 has created a situation of global emergency all over the world from the last few months. We are witnessing a helpless situation due to COVID-19 as no vaccine or drug is effective against the disease. In the present study, we have tested the repurposing efficacy of some currently used combination drugs against COVID-19. We have tried to understand the mechanism of action of some repurposed drugs:Favipiravir (F), Hydroxychloroquine (H) and Oseltamivir (O). The ADME analysis have suggested strong inhibitory possibility of F, H, O combination towards receptor protein of 3CL<jats:sup>pro</jats:sup> of SARS-CoV-2 virus. The strong binding affinity, number of hydrogen bond interaction between inhibitor, receptor and lower inhibition constant computed from molecular docking validated the better complexation possibility of F + H + O:3CL<jats:sup>pro</jats:sup>combination. Various thermodynamical output from Molecular dynamics (MD) simulations like potential energy (E<jats:sub>g</jats:sub>), temperature (T), density, pressure, SASA energy, interaction energies, Gibbs free energy (ΔG<jats:sub>bind</jats:sub>) etc., also favored the complexation between F + H + O and CoV-2 protease. Our in-silico results have recommended the strong candidature of combination drugs Favipiravir, Hydroxychloroquine and Oseltamivir as a potential lead inhibitor for targeting SARS-CoV-2 infections.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
6,
16
]
]
},
"author": [
{
"affiliation": [
{
"name": "Jaypee Institute of Information Technology"
}
],
"family": "Yadav",
"given": "Pooja",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-9702-2553",
"affiliation": [
{
"name": "Jaypee Institute of Information Technology"
}
],
"authenticated-orcid": false,
"family": "CHOWDHURY",
"given": "PAPIA",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
6,
21
]
],
"date-time": "2021-06-21T15:19:25Z",
"timestamp": 1624288765000
},
"deposited": {
"date-parts": [
[
2021,
6,
21
]
],
"date-time": "2021-06-21T15:19:27Z",
"timestamp": 1624288767000
},
"group-title": "In Review",
"indexed": {
"date-parts": [
[
2022,
4,
2
]
],
"date-time": "2022-04-02T18:29:46Z",
"timestamp": 1648924186483
},
"institution": [
{
"name": "Research Square"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2021,
6,
21
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
6,
21
]
],
"date-time": "2021-06-21T00:00:00Z",
"timestamp": 1624233600000
}
}
],
"link": [
{
"URL": "https://www.researchsquare.com/article/rs-628277/v1",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.researchsquare.com/article/rs-628277/v1.html",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "8761",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
6,
21
]
]
},
"prefix": "10.21203",
"published": {
"date-parts": [
[
2021,
6,
21
]
]
},
"publisher": "Research Square Platform LLC",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.researchsquare.com/article/rs-628277/v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study",
"type": "posted-content"
}