Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein
et al., bioRxiv, doi:10.1101/2021.03.16.435741, Mar 2021
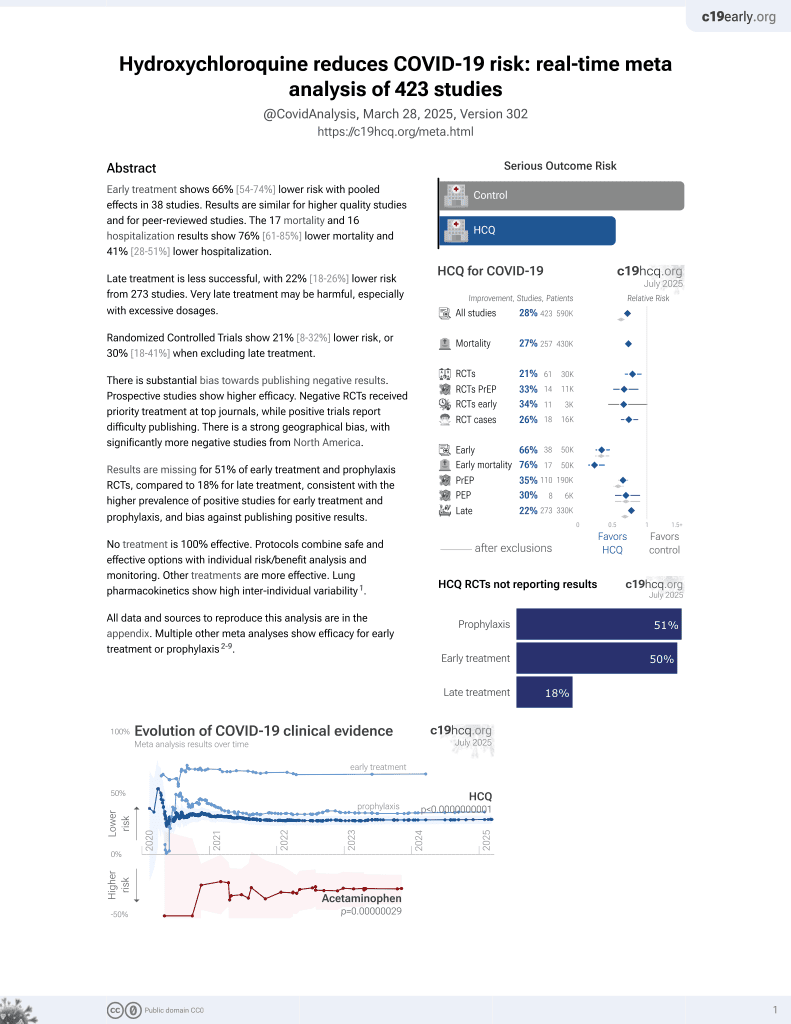
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 424 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Microscopy/spectroscopy study showing that HCQ binds to both N-terminal domain and C-terminal domain of SARS-CoV-2 nucleocapsid protein to inhibit their interactions with nucleic acids and disrupt NA-induced liquid-liquid phase separation essential for the viral life cycle including the package of gRNA and N protein into new virions. These results suggest that HCQ may achieve its anti-SARS-CoV-2 activity by interfering in several key steps of the viral life cycle.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Dang et al., 17 Mar 2021, preprint, 2 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Structural basis of anti-SARS-CoV-2 activity of HCQ: specific binding to N protein to disrupt its interaction with nucleic acids and LLPS
doi:10.1101/2021.03.16.435741
Great efforts have led to successfully developing the spike-based vaccines but challenges still exist to completely terminate the SARS-CoV-2 pandemic. SARS-CoV-2 nucleocapsid (N) protein plays the essential roles in almost all key steps of the viral life cycle, thus representing a top drug target. Almost all key functions of N protein including liquid-liquid phase separation (LLPS) depend on its capacity in interacting with nucleic acids. Therefore, only the variants with their N proteins functional in binding nucleic acids might survive and spread in evolution and indeed, the residues critical for binding nucleic acids are highly conserved. Very recently, hydroxychloroquine (HCQ) was shown to prevent the transmission in a large-scale clinical study in Singapore but so far, no specific SARS-CoV-2 protein was experimentally identified to be targeted by HCQ. Here by NMR, we unambiguously decode that HCQ specifically binds NTD and CTD of SARS-CoV-2 N protein with Kd of 112.1 and 57.1 µM respectively to inhibit their interaction with nucleic acid, as well as to disrupt LLPS essential for the viral life cycle. Most importantly, HCQ-binding residues are identical in SARS-CoV-2 variants and therefore HCQ is likely effective to them all. The results not only provide a structural basis for the anti-SARS-CoV-2 activity of HCQ, but also renders HCQ to be the first known drug capable of targeting LLPS. Furthermore, the unique structure of the HCQ-CTD complex decodes a promising strategy for further design of better anti-SARS-CoV-2 drugs from HCQ. Therefore, HCQ is a promising candidate to help terminate the pandemic.
Molecular docking The structures of the HCQ-NTD and HCQ-CTD complex were constructed by use of the well-established HADDOCK software (10, 20, 22, 23) in combination with crystallography and NMR system (CNS) (38) , which makes use of CSD data to derive the docking that allows various degrees of flexibility. Briefly, HADDOCK docking was performed in three stages: (1) randomization and rigid body docking; (2) semi-flexible simulated annealing; and (3) flexible explicit solvent refinement. The NMR structure (2) of NTD (PDB ID of 6YI3) and crystal structure (3) of CTD (PDB ID of 6YUN) were used for docking to HCQ. The HCQ-NTD and HCQ-CTD structures with the lowest energy score were selected for the detailed analysis and display by Pymol (The PyMOL Molecular Graphics System).
Author contributions Conceived the research: J.S. Performed research and analyzed data: M.D and J.S; Acquired funding: J.S; Wrote manuscript: J.S.
Supplementary Materials
Table S1 Figure S1-S7
References
Brunger, Crystallography & NMR system: a new software suite for macromolecular structure determination, Acta Crystallogr. Sect. D
Carlson, Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions, Mol Cell
Chen, Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA, J Mol Biol
Dang, ATP biphasically modulates LLPS of SARS-CoV-2 nucleocapsid protein and specifically binds its RNA-binding domain, Biochem Biophys Res Commun
Delaglio, NMRPipe: A multidimensional spectral processing system based on UNIX pipes, J Biomol NMR
Dinesh, Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein, PLoS Pathog
Dominguez, HADDOCK: a protein-protein docking approach based on biochemical or biophysical information, J. Am. Chem. Soc
Hoepel, High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages, Sci Transl Med doi
Hyman, Liquid-liquid phase separation in biology, Annu Rev Cell Dev Biol
Iserman, Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2
Johnson, Blevins, NMRView: A computer program for the visualization and analysis of NMR data, J. Biomol. NMR
Kang, A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids, PLoS Biol
Kang, ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain, Commun Biol
Korn, H, (13)C, and (15)N backbone chemical shift assignments of the Cterminal dimerization domain of SARS-CoV-2 nucleocapsid protein, Biomol NMR Assign
Lee, Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies, Nat Microbiol
Lei, SARS-CoV-2 Spike Protein Impairs Endothelial Function via Downregulation of ACE 2, Circ Res
Lim, Curcumin Allosterically Inhibits the Dengue NS2B-NS3 Protease by
Liu, An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies, Cell, doi:10.1016/j.cell.2021.05.032
Lu, The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein, Nat Commun
Pace, How to measure and predict the molar absorption coefficient of a protein, Protein Sci
Patel, ATP as a biological hydrotrope, Science
Perdikari, SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs, EMBO J
Qin, Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor, J Biol Chem
Qin, Dynamic principle for designing antagonistic/agonistic molecules for EphA4 receptor, the only known ALS modifier, ACS Chem Biol
Roldan, The possible mechanisms of action of 4-aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): A role for iron homeostasis?, Pharmacol Res
Satarker, Hydroxychloroquine in COVID-19: Potential Mechanism of Action Against SARS-CoV-2
Sattler, Heteronuclear multidimensional NMR experiments for the struc-ture determination of proteins in solution employing pulsed field gradients, Prog. NMR Spectroscopy
Savastano, Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates, Nat Commun
Seet, Positive impact of oral hydroxychloroquine and povidone-iodine throat spray for COVID-19 prophylaxis: An open-label randomized trial, Int J Infect Dis
Shi, Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-Like protease by the N214A mutation on the extra domain, PLoS Comput Biol
Shin, Brangwynne, Liquid phase condensation in cell physiology and disease, Science
Song, Adenosine triphosphate energy-independently controls protein homeostasis with unique structure and diverse mechanisms, Protein Sci. Apr, doi:10.1002/pro.4079
Song, Ni, NMR for the design of functional mimetics of protein-protein interactions: one key is in the building of bridges, Biochem. Cell Biol
Wang, Antibody Resistance of SARS-CoV-2 Variants B, Nature. Mar, doi:10.1038/s41586-021-03398-2
Williamson, Using chemical shift perturbation to characterise ligand binding, Prog. Nucl. Magn. Reson. Spectrosc
Wu, new coronavirus associated with human respiratory disease in China, Nature
Zhang, Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues, Proc Natl Acad Sci U S A
Zinzula, High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2, Biochem Biophys Res Commun
DOI record:
{
"DOI": "10.1101/2021.03.16.435741",
"URL": "http://dx.doi.org/10.1101/2021.03.16.435741",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Great efforts have led to successfully developing the spike-based vaccines but challenges still exist to completely terminate the SARS-CoV-2 pandemic. SARS-CoV-2 nucleocapsid (N) protein plays the essential roles in almost all key steps of the viral life cycle, thus representing a top drug target. Almost all key functions of N protein including liquid-liquid phase separation (LLPS) depend on its capacity in interacting with nucleic acids. Therefore, only the variants with their N proteins functional in binding nucleic acids might survive and spread in evolution and indeed, the residues critical for binding nucleic acids are highly conserved. Very recently, hydroxychloroquine (HCQ) was shown to prevent the transmission in a large-scale clinical study in Singapore but so far, no specific SARS-CoV-2 protein was experimentally identified to be targeted by HCQ. Here by NMR, we unambiguously decode that HCQ specifically binds NTD and CTD of SARS-CoV-2 N protein with Kd of 112.1 and 57.1 μM respectively to inhibit their interaction with nucleic acid, as well as to disrupt LLPS essential for the viral life cycle. Most importantly, HCQ-binding residues are identical in SARS-CoV-2 variants and therefore HCQ is likely effective to them all. The results not only provide a structural basis for the anti-SARS-CoV-2 activity of HCQ, but also renders HCQ to be the first known drug capable of targeting LLPS. Furthermore, the unique structure of the HCQ-CTD complex decodes a promising strategy for further design of better anti-SARS-CoV-2 drugs from HCQ. Therefore, HCQ is a promising candidate to help terminate the pandemic.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
6,
15
]
]
},
"author": [
{
"affiliation": [],
"family": "Dang",
"given": "Mei",
"sequence": "first"
},
{
"affiliation": [],
"family": "Song",
"given": "Jianxing",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
3,
18
]
],
"date-time": "2021-03-18T04:20:14Z",
"timestamp": 1616041214000
},
"deposited": {
"date-parts": [
[
2022,
5,
25
]
],
"date-time": "2022-05-25T19:44:47Z",
"timestamp": 1653507887000
},
"group-title": "Biophysics",
"indexed": {
"date-parts": [
[
2024,
3,
1
]
],
"date-time": "2024-03-01T23:42:31Z",
"timestamp": 1709336551608
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 1,
"issued": {
"date-parts": [
[
2021,
3,
17
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2021.03.16.435741",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
3,
17
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2021,
3,
17
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.1"
},
{
"DOI": "10.1371/journal.ppat.1009100",
"article-title": "Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein",
"doi-asserted-by": "crossref",
"first-page": "e1009100",
"journal-title": "PLoS Pathog",
"key": "2021061705151168000_2021.03.16.435741v3.2",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2020.09.131",
"article-title": "High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2",
"doi-asserted-by": "crossref",
"first-page": "54",
"journal-title": "Biochem Biophys Res Commun",
"key": "2021061705151168000_2021.03.16.435741v3.3",
"volume": "538",
"year": "2021"
},
{
"DOI": "10.1016/j.jmb.2007.02.069",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.4"
},
{
"DOI": "10.1038/s41467-020-20768-y",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.5"
},
{
"DOI": "10.1038/s41467-020-19843-1",
"article-title": "Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates",
"doi-asserted-by": "crossref",
"first-page": "6041",
"journal-title": "Nat Commun",
"key": "2021061705151168000_2021.03.16.435741v3.6",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2020.11.041",
"article-title": "Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid",
"doi-asserted-by": "crossref",
"first-page": "1078",
"journal-title": "Mol Cell",
"key": "2021061705151168000_2021.03.16.435741v3.7",
"volume": "80",
"year": "2020"
},
{
"article-title": "SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs",
"first-page": "e106478",
"journal-title": "EMBO J",
"key": "2021061705151168000_2021.03.16.435741v3.8",
"volume": "17",
"year": "2020"
},
{
"article-title": "Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions",
"first-page": "1092",
"journal-title": "Mol Cell",
"key": "2021061705151168000_2021.03.16.435741v3.9",
"volume": "17",
"year": "2020"
},
{
"first-page": "50",
"journal-title": "ATP biphasically modulates LLPS of SARS-CoV-2 nucleocapsid protein and specifically binds its RNA-binding domain Biochem Biophys Res Commun",
"key": "2021061705151168000_2021.03.16.435741v3.10",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1146/annurev-cellbio-100913-013325",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.11"
},
{
"DOI": "10.1126/science.aaf4382",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.12"
},
{
"DOI": "10.1126/science.aaf6846",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.13"
},
{
"DOI": "10.1371/journal.pbio.3000327",
"article-title": "A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids",
"doi-asserted-by": "crossref",
"first-page": "e3000327",
"journal-title": "PLoS Biol",
"key": "2021061705151168000_2021.03.16.435741v3.14",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1002/pro.4079",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.15"
},
{
"DOI": "10.1016/j.phrs.2020.104904",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.16"
},
{
"first-page": "1",
"journal-title": "Hydroxychloroquine in COVID-19: Potential Mechanism of Action Against SARS-CoV-2 Curr Pharmacol Rep",
"key": "2021061705151168000_2021.03.16.435741v3.17",
"volume": "24",
"year": "2020"
},
{
"DOI": "10.1016/j.ijid.2021.04.035",
"article-title": "Positive impact of oral hydroxychloroquine and povidone-iodine throat spray for COVID-19 prophylaxis: An open-label randomized trial",
"doi-asserted-by": "crossref",
"first-page": "314",
"journal-title": "Int J Infect Dis",
"key": "2021061705151168000_2021.03.16.435741v3.18",
"volume": "106",
"year": "2021"
},
{
"DOI": "10.1016/j.pnmrs.2013.02.001",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.19"
},
{
"DOI": "10.1074/jbc.M804114200",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.20"
},
{
"DOI": "10.1021/cb500413n",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.21"
},
{
"article-title": "ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain",
"first-page": "223",
"issue": "2",
"journal-title": "Commun Biol",
"key": "2021061705151168000_2021.03.16.435741v3.22",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1021/ja026939x",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.23"
},
{
"DOI": "10.1038/s41586-021-03398-2",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.24"
},
{
"DOI": "10.1161/CIRCRESAHA.121.318902",
"article-title": "SARS-CoV-2 Spike Protein Impairs Endothelial Function via Downregulation of ACE 2",
"doi-asserted-by": "crossref",
"first-page": "1323",
"journal-title": "Circ Res",
"key": "2021061705151168000_2021.03.16.435741v3.25",
"volume": "128",
"year": "2021"
},
{
"DOI": "10.1126/scitranslmed.abf8654",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.26"
},
{
"article-title": "Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies",
"first-page": "1185",
"journal-title": "Nat Microbiol",
"key": "2021061705151168000_2021.03.16.435741v3.27",
"volume": "5",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.05.032",
"doi-asserted-by": "crossref",
"key": "2021061705151168000_2021.03.16.435741v3.28",
"unstructured": "Y. Liu et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. https://doi.org/10.1016/j.cell.2021.05.032 (2021)."
},
{
"DOI": "10.1073/pnas.2105968118",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.29"
},
{
"DOI": "10.1139/bcb-76-2-3-177",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.30"
},
{
"DOI": "10.1371/journal.pcbi.1001084",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.31"
},
{
"DOI": "10.1021/acsomega.0c00039",
"article-title": "Curcumin Allosterically Inhibits the Dengue NS2B-NS3 Protease by Disrupting Its Active Conformation",
"doi-asserted-by": "crossref",
"first-page": "25677",
"journal-title": "ACS Omega",
"key": "2021061705151168000_2021.03.16.435741v3.32",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1002/pro.5560041120",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.33"
},
{
"DOI": "10.1002/(SICI)1521-3749(199901)625:1<93::AID-ZAAC93>3.0.CO;2-4",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.34"
},
{
"DOI": "10.1007/BF00197809",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.35"
},
{
"DOI": "10.1007/BF00404272",
"doi-asserted-by": "publisher",
"key": "2021061705151168000_2021.03.16.435741v3.36"
},
{
"DOI": "10.1007/s12104-020-09995-y",
"article-title": "(1)H, (13)C, and (15)N backbone chemical shift assignments of the C-terminal dimerization domain of SARS-CoV-2 nucleocapsid protein",
"doi-asserted-by": "crossref",
"first-page": "129",
"journal-title": "Biomol NMR Assign",
"key": "2021061705151168000_2021.03.16.435741v3.37",
"volume": "15",
"year": "2021"
},
{
"article-title": "Crystallography & NMR system: a new software suite for macromolecular structure determination",
"first-page": "905",
"journal-title": "Acta Crystallogr. Sect",
"key": "2021061705151168000_2021.03.16.435741v3.38",
"volume": "D 54",
"year": "1998"
}
],
"reference-count": 38,
"references-count": 38,
"relation": {
"is-preprint-of": [
{
"asserted-by": "subject",
"id": "10.1017/qrd.2021.12",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "http://biorxiv.org/lookup/doi/10.1101/2021.03.16.435741"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Structural basis of anti-SARS-CoV-2 activity of HCQ: specific binding to N protein to disrupt its interaction with nucleic acids and LLPS",
"type": "posted-content"
}