Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection
et al., ChemBioChem, doi:10.1002/cbic.202400404, Jun 2024
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 424 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
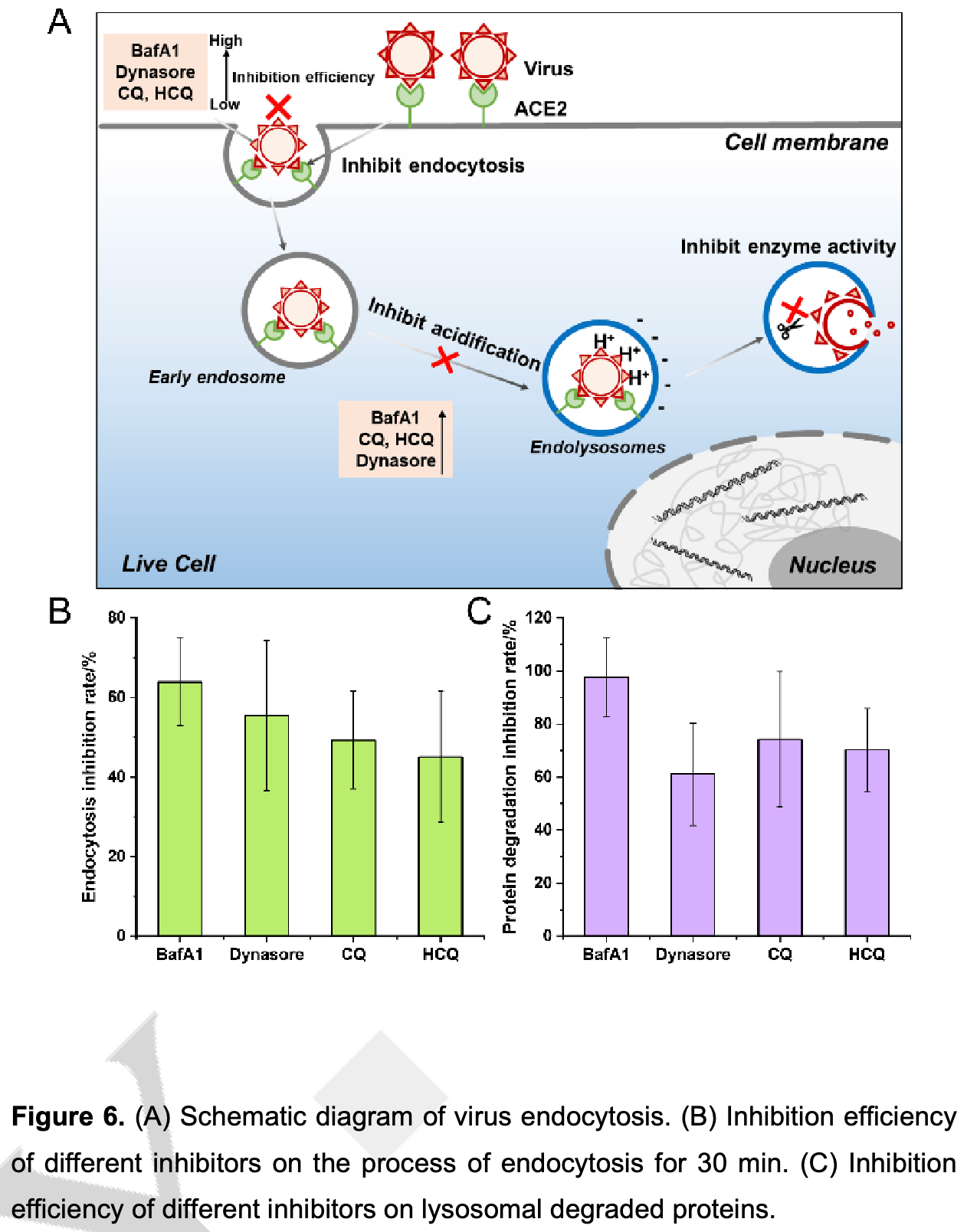
In vitro study showing that endosomal acidification inhibitors like HCQ minimize SARS-CoV-2 infection by blocking viral internalization and RNA release in cells expressing ACE2. Authors utilized super-resolution structured illumination microscopy (SIM) to analyze the effects of CQ, HCQ, Bafilomycin A1 (BafA1), and Dynasore on the endocytic processes of SARS-CoV-2. All four compounds inhibited the internalization and degradation of the RBD-ACE2 complex in living cells, with BafA1 showing the highest inhibition rate. The results confirm that endosomal acidification inhibitors can disrupt critical stages of SARS-CoV-2 infection within host cells.
See also Shang et al., an in vitro and mouse study showing that endosomal acidification inhibitors CQ, bafilomycin A1, and NH4CL suppress SARS-CoV-2 replication and relieve viral pneumonia. Authors found that these compounds significantly reduced SARS-CoV-2 viral yields in Vero E6, Huh-7, and 293T-ACE2 cells. In hACE2 transgenic mice, CQ and bafilomycin A1 reduced viral replication in lung tissues and alleviated pneumonia with reduced inflammatory infiltration and improved alveolar structure.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
2.
Shang (B) et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
3.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
4.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
5.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
6.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
8.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
9.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
10.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
11.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
12.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
13.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
14.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
15.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
16.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
17.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
18.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
19.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
20.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
21.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
22.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
23.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
24.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
25.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
26.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
27.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Yan et al., 14 Jun 2024, peer-reviewed, 7 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
DOI record:
{
"DOI": "10.1002/cbic.202400404",
"ISSN": [
"1439-4227",
"1439-7633"
],
"URL": "http://dx.doi.org/10.1002/cbic.202400404",
"abstract": "<jats:p>In this study, super‐resolution structured illumination microscope (SIM) was used to analyze molecular mechanism of endocytic acidification inhibitors in the SARS‐CoV‐2 pandemic, such as Chloroquine (CQ), Hydroxychloroquine (HCQ) and Bafilomycin A1 (BafA1). We fluorescently labeled the SARS‐CoV‐2 RBD and its receptor ACE2 protein with small molecule dyes. Utilizing SIM imaging, the real‐time impact of inhibitors (BafA1, CQ, HCQ, Dynasore) on the RBD‐ACE2 endocytotic process was dynamically tracked in living cells. Initially, the protein activity of RBD and ACE2 was ensured after being labeled. And then our findings revealed that these inhibitors could inhibit the internalization and degradation of RBD‐ACE2 to varying degrees. Among them, 100 nM BafA1 exhibited the most satisfactory endocytotic inhibition (~63.9%) and protein degradation inhibition (~97.7%). And it could inhibit the fusion between endocytic vesicles in the living cells. Additionally, Dynasore, a widely recognized dynein inhibitor, also demonstrated cell acidification inhibition effects. Together, these inhibitors collectively hinder SARS‐CoV‐2 infection by inhibiting both the viral internalization and RNA release. The comprehensive evaluation of pharmacological mechanisms through super‐resolution fluorescence imaging has laid a crucial theoretical foundation for the development of potential drugs to treat COVID‐19.</jats:p>",
"alternative-id": [
"10.1002/cbic.202400404"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-05-02"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2024-06-14"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2024-06-14"
}
],
"author": [
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Yan",
"given": "Chunyu",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Zhou",
"given": "Wei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Zhang",
"given": "Yan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Zhou",
"given": "Xuelian",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Qiao",
"given": "Qinglong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics department of biotechnology CHINA"
}
],
"family": "Miao",
"given": "Lu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dalian Institute of Chemical Physics Department of Biotechnology Department of Biological Technology 457 Zhongshan Road 116023 Dalian CHINA"
}
],
"family": "Xu",
"given": "Zhaochao",
"sequence": "additional"
}
],
"container-title": "ChemBioChem",
"container-title-short": "ChemBioChem",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"chemistry-europe.onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2024,
6,
15
]
],
"date-time": "2024-06-15T03:10:25Z",
"timestamp": 1718421025000
},
"deposited": {
"date-parts": [
[
2024,
6,
15
]
],
"date-time": "2024-06-15T03:10:27Z",
"timestamp": 1718421027000
},
"indexed": {
"date-parts": [
[
2024,
6,
16
]
],
"date-time": "2024-06-16T00:19:30Z",
"timestamp": 1718497170381
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
6,
14
]
]
},
"language": "en",
"license": [
{
"URL": "http://onlinelibrary.wiley.com/termsAndConditions#vor",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
6,
14
]
],
"date-time": "2024-06-14T00:00:00Z",
"timestamp": 1718323200000
}
}
],
"member": "311",
"original-title": [],
"prefix": "10.1002",
"published": {
"date-parts": [
[
2024,
6,
14
]
]
},
"published-online": {
"date-parts": [
[
2024,
6,
14
]
]
},
"publisher": "Wiley",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cbic.202400404"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Super‐resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS‐CoV‐2 infection",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy"
}