Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics
et al., Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314, Oct 2025
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 423 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
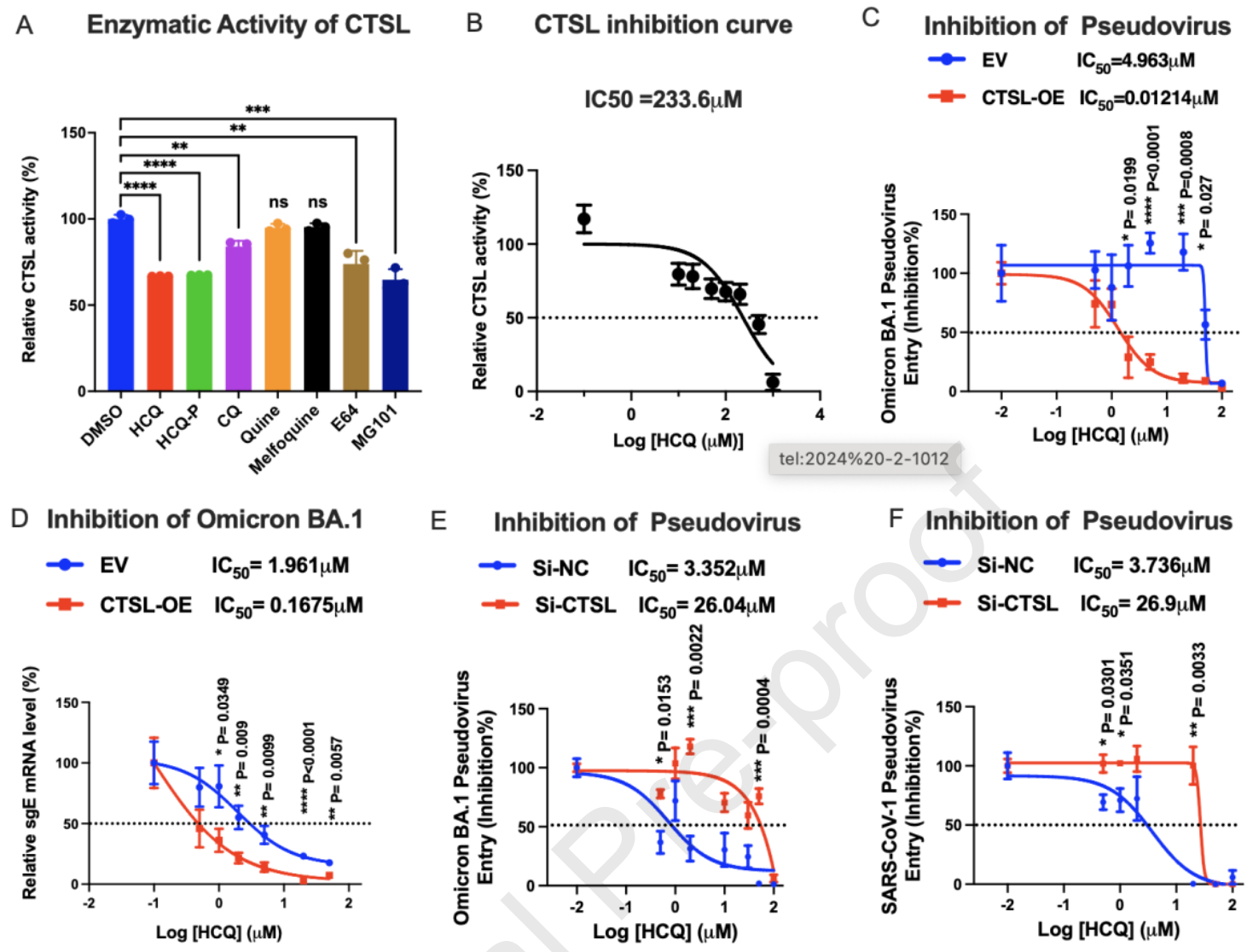
In vitro study showing that hydroxychloroquine (HCQ) inhibits SARS-CoV-2 variants through direct binding to cathepsin L (CTSL), with enhanced efficacy against Omicron BA.1 compared to Delta in VeroE6 and Huh7 cells. Authors developed a clickable photo-crosslinking probe (HCQ-P) combined with quantitative proteomics to identify CTSL as HCQ's primary molecular target. HCQ significantly inhibited CTSL protease activity at 50μM and suppressed CTSL-dependent coronavirus entry into cells that utilize the endosomal pathway rather than TMPRSS2-mediated entry. In Huh7 cells overexpressing CTSL, HCQ showed 408-fold lower IC50 (0.01241μM) compared to control cells (4.963μM) against Omicron BA.1 pseudovirus infection. Authors confirmed HCQ's antiviral mechanism through molecular docking studies showing HCQ binding to CTSL's hydrophobic pocket involving residues Cys26, Gly69, and Ser217. CTSL knockdown experiments demonstrated that HCQ's antiviral efficacy was dramatically reduced when CTSL was depleted, confirming CTSL-dependence.
Authors note that CTSL is a key, but not the only target of HCQ - the multiple functions of HCQ suggest that it may engage several targets and the mechanisms may work synergistically.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Shang et al., 18 Oct 2025, China, peer-reviewed, 8 authors.
Contact: q.zhao@polyu.edu.hk, hinchu@hku.hk, mankin.wong@polyu.edu.hk.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: Journal Pre-proof
Identification of Cathepsin L as the molecular target of hydroxychloroquine with
chemical proteomics
Jin Shang, Bingjie Hu, Ka-Yan Karen Kung, Jiajun Jiang, Qi Zhang, Man-Kin Wong,
Hin Chu, Qian Zhao
PII:
S1535-9476(25)00413-X
DOI:
https://doi.org/10.1016/j.mcpro.2025.101314
Reference:
MCPRO 101314
To appear in:
Molecular & Cellular Proteomics
Received Date: 22 April 2025
Revised Date:
21 September 2025
Accepted Date: 15 October 2025
Please cite this article as: Shang J, Hu B, Kung KYK, Jiang J, Zhang Q, Wong MK, Chu H, Zhao Q,
Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics,
Molecular & Cellular Proteomics (2025), doi: https://doi.org/10.1016/j.mcpro.2025.101314.
This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition
of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of
record. This version will undergo additional copyediting, typesetting and review before it is published
in its final form, but we are providing this version to give early visibility of the article. Please note that,
during the production process, errors may be discovered which could affect the content, and all legal
disclaimers that apply to the journal pertain.
® 2025 THE AUTHORS. Published by Elsevier Inc on behalf of American Society for Biochemistry and
Molecular Biology.
ur
na
Jo
lP
re
-p
r
oo
f
Identification of Cathepsin L as the molecular target of
hydroxychloroquine with chemical proteomics.
Jin Shang1, 2#, Bingjie Hu3#, Ka-Yan Karen Kung1, 3, Jiajun Jiang1, 4, Qi Zhang1, ManKin Wong1, 4*, Hin Chu3*, Qian Zhao1,2*
1
State Key Laboratory of Chemical Biology and Drug Discovery, Department of Applied Biological
oo
f
and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Hong Kong SAR,
China.
Department of Applied Biological and Chemical Technology, The Hong Kong Polytechnic
re
-p
r
2
University, Hung Hom, Hong Kong SAR, China.
3
State Key Laboratory of Emerging Infectious Diseases, Department of Microbiology, School of
Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong
4
lP
Kong SAR, China.
Department of Food Science and Nutrition, The Hong Kong Polytechnic University, Hung Hom,
Hong Kong SAR, China
The authors contributed equally
*
Corresponding Author: Qian ZHAO (q.zhao@polyu.edu.hk); Hin Chu (hinchu@hku.hk); Man-Kin
ur
na
#
Wong (mankin.wong@polyu.edu.hk).
Jo
ABSTRACT: Hydroxychloroquine (HCQ) and chloroquine (CQ) have been utilized as antimalarial drugs for decades. Recently, these compounds were reported to inhibit various viruses
utilizing the endosomal entry pathway. However, their direct molecular targets in host cells
remain elusive. In this study, we developed a clickable photo-crosslinking probe in combination
with proteomic approaches to identified Cathepsin L (CTSL) as the binding target of HCQ.
Extensive biochemical and in silico analyses were conducted to validate the HCQ-CTSL
interactions. HCQ significantly inhibited the protease activity of CTSL and suppressed CTSLdependent coronavirus entry in cells that support endosomal entry pathway. These findings not
only reveal the underlying mechanism of how HCQ inhibits endosomal viral entry but also
guide the rational use of HCQ against other emerging infectious agents.
DOI record:
{
"DOI": "10.1016/j.mcpro.2025.101314",
"ISSN": [
"1535-9476"
],
"URL": "http://dx.doi.org/10.1016/j.mcpro.2025.101314",
"alternative-id": [
"S153594762500413X"
],
"article-number": "101314",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Molecular & Cellular Proteomics"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.mcpro.2025.101314"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "® 2025 THE AUTHORS. Published by Elsevier Inc on behalf of American Society for Biochemistry and Molecular Biology."
}
],
"author": [
{
"affiliation": [],
"family": "Shang",
"given": "Jin",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-6677-9454",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hu",
"given": "Bingjie",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-2732-4369",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kung",
"given": "Ka-Yan Karen",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jiang",
"given": "Jiajun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Qi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wong",
"given": "Man-Kin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chu",
"given": "Hin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2244-6516",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhao",
"given": "Qian",
"sequence": "additional"
}
],
"container-title": "Molecular & Cellular Proteomics",
"container-title-short": "Molecular & Cellular Proteomics",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"mcponline.org",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
10,
18
]
],
"date-time": "2025-10-18T22:39:20Z",
"timestamp": 1760827160000
},
"deposited": {
"date-parts": [
[
2025,
10,
18
]
],
"date-time": "2025-10-18T22:39:24Z",
"timestamp": 1760827164000
},
"indexed": {
"date-parts": [
[
2025,
10,
18
]
],
"date-time": "2025-10-18T23:10:30Z",
"timestamp": 1760829030139,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
10
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
1
]
],
"date-time": "2025-10-01T00:00:00Z",
"timestamp": 1759276800000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
1
]
],
"date-time": "2025-10-01T00:00:00Z",
"timestamp": 1759276800000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 16,
"start": {
"date-parts": [
[
2025,
10,
17
]
],
"date-time": "2025-10-17T00:00:00Z",
"timestamp": 1760659200000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S153594762500413X?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S153594762500413X?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "101314",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2025,
10
]
]
},
"published-print": {
"date-parts": [
[
2025,
10
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1021/jm990437l",
"article-title": "Structure− function relationships in aminoquinolines: effect of amino and chloro groups on quinoline− hematin complex formation, inhibition of β-hematin formation, and antiplasmodial activity",
"author": "Egan",
"doi-asserted-by": "crossref",
"first-page": "283",
"issue": "2",
"journal-title": "Journal of medicinal chemistry",
"key": "10.1016/j.mcpro.2025.101314_bib1",
"volume": "43",
"year": "2000"
},
{
"DOI": "10.1016/j.ebiom.2017.09.034",
"article-title": "Chloroquine, a FDA-approved drug, prevents Zika virus infection and its associated congenital microcephaly in mice",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "189",
"journal-title": "EBioMedicine",
"key": "10.1016/j.mcpro.2025.101314_bib2",
"volume": "24",
"year": "2017"
},
{
"DOI": "10.1158/2159-8290.CD-18-0706",
"article-title": "PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer",
"author": "Rebecca",
"doi-asserted-by": "crossref",
"first-page": "220",
"issue": "2",
"journal-title": "Cancer discovery",
"key": "10.1016/j.mcpro.2025.101314_bib3",
"volume": "9",
"year": "2019"
},
{
"DOI": "10.1021/acscentsci.2c01325",
"article-title": "A turn-on fluorescent amino acid sensor reveals chloroquine’s effect on cellular amino acids via inhibiting Cathepsin L",
"author": "Smith",
"doi-asserted-by": "crossref",
"first-page": "980",
"issue": "5",
"journal-title": "ACS Central Science",
"key": "10.1016/j.mcpro.2025.101314_bib4",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1128/AAC.01509-08",
"article-title": "Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice",
"author": "Keyaerts",
"doi-asserted-by": "crossref",
"first-page": "3416",
"issue": "8",
"journal-title": "Antimicrobial agents and chemotherapy",
"key": "10.1016/j.mcpro.2025.101314_bib5",
"volume": "53",
"year": "2009"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105938",
"article-title": "New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?",
"author": "Devaux",
"doi-asserted-by": "crossref",
"issue": "5",
"journal-title": "International journal of antimicrobial agents",
"key": "10.1016/j.mcpro.2025.101314_bib6",
"volume": "55",
"year": "2020"
},
{
"DOI": "10.1016/S1473-3099(03)00806-5",
"article-title": "Effects of chloroquine on viral infections: an old drug against today's diseases",
"author": "Savarino",
"doi-asserted-by": "crossref",
"first-page": "722",
"issue": "11",
"journal-title": "The Lancet infectious diseases",
"key": "10.1016/j.mcpro.2025.101314_bib7",
"volume": "3",
"year": "2003"
},
{
"DOI": "10.1016/j.tmaid.2020.101735",
"article-title": "Therapeutic use of chloroquine and hydroxychloroquine in COVID-19 and other viral infections: A narrative review",
"author": "Hashem",
"doi-asserted-by": "crossref",
"journal-title": "Travel medicine and infectious disease",
"key": "10.1016/j.mcpro.2025.101314_bib8",
"volume": "35",
"year": "2020"
},
{
"DOI": "10.1002/j.1460-2075.1984.tb02195.x",
"article-title": "Identification of the acidic compartment of Plasmodium falciparum‐infected human erythrocytes as the target of the antimalarial drug chloroquine",
"author": "Yayon",
"doi-asserted-by": "crossref",
"first-page": "2695",
"issue": "11",
"journal-title": "The EMBO journal",
"key": "10.1016/j.mcpro.2025.101314_bib9",
"volume": "3",
"year": "1984"
},
{
"DOI": "10.1016/0163-7258(93)90056-J",
"article-title": "Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum",
"author": "Slater",
"doi-asserted-by": "crossref",
"first-page": "203",
"issue": "2-3",
"journal-title": "Pharmacology & therapeutics",
"key": "10.1016/j.mcpro.2025.101314_bib10",
"volume": "57",
"year": "1993"
},
{
"DOI": "10.1038/s41584-020-0372-x",
"article-title": "Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology",
"author": "Schrezenmeier",
"doi-asserted-by": "crossref",
"first-page": "155",
"issue": "3",
"journal-title": "Nature Reviews Rheumatology",
"key": "10.1016/j.mcpro.2025.101314_bib11",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"issue": "2",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101314_bib12",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-20457-w",
"article-title": "Host and viral determinants for efficient SARS-CoV-2 infection of the human lung",
"author": "Chu",
"doi-asserted-by": "crossref",
"first-page": "134",
"issue": "1",
"journal-title": "Nat Commun",
"key": "10.1016/j.mcpro.2025.101314_bib13",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.09.033",
"article-title": "SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2",
"author": "Clausen",
"doi-asserted-by": "crossref",
"first-page": "1043",
"issue": "4",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101314_bib14",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1016/j.jbc.2021.100759",
"article-title": "The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection",
"author": "Carlos",
"doi-asserted-by": "crossref",
"journal-title": "J Biol Chem",
"key": "10.1016/j.mcpro.2025.101314_bib15",
"volume": "296",
"year": "2021"
},
{
"DOI": "10.1126/science.abd3072",
"article-title": "Neuropilin-1 is a host factor for SARS-CoV-2 infection",
"author": "Daly",
"doi-asserted-by": "crossref",
"first-page": "861",
"issue": "6518",
"journal-title": "Science",
"key": "10.1016/j.mcpro.2025.101314_bib16",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1038/s42255-020-00324-0",
"article-title": "HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "1391",
"issue": "12",
"journal-title": "Nat Metab",
"key": "10.1016/j.mcpro.2025.101314_bib17",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1021/acscentsci.0c01537",
"article-title": "CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2",
"author": "Amraei",
"doi-asserted-by": "crossref",
"first-page": "1156",
"issue": "7",
"journal-title": "ACS Cent Sci",
"key": "10.1016/j.mcpro.2025.101314_bib18",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1038/s41422-020-00460-y",
"article-title": "AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "126",
"issue": "2",
"journal-title": "Cell Res",
"key": "10.1016/j.mcpro.2025.101314_bib19",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2023.06.005",
"article-title": "TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry",
"author": "Baggen",
"doi-asserted-by": "crossref",
"first-page": "3427",
"issue": "16",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101314_bib20",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1016/j.molcel.2020.04.022",
"article-title": "A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "779",
"issue": "4",
"journal-title": "Mol Cell",
"key": "10.1016/j.mcpro.2025.101314_bib21",
"volume": "78",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-25729-7",
"article-title": "A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses",
"author": "Beumer",
"doi-asserted-by": "crossref",
"first-page": "5498",
"issue": "1",
"journal-title": "Nat Commun",
"key": "10.1016/j.mcpro.2025.101314_bib22",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1126/sciadv.add3867",
"article-title": "Altered host protease determinants for SARS-CoV-2 Omicron",
"author": "Chan",
"doi-asserted-by": "crossref",
"first-page": "eadd3867",
"issue": "3",
"journal-title": "Sci Adv",
"key": "10.1016/j.mcpro.2025.101314_bib23",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1128/jvi.00851-23",
"article-title": "SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models",
"author": "Mykytyn",
"doi-asserted-by": "crossref",
"first-page": "e0085123",
"issue": "8",
"journal-title": "J Virol",
"key": "10.1016/j.mcpro.2025.101314_bib24",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1016/j.ebiom.2021.103255",
"article-title": "Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"journal-title": "EBioMedicine",
"key": "10.1016/j.mcpro.2025.101314_bib25",
"volume": "65",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2575-3",
"article-title": "Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "588",
"issue": "7826",
"journal-title": "Nature",
"key": "10.1016/j.mcpro.2025.101314_bib26",
"volume": "585",
"year": "2020"
},
{
"DOI": "10.15252/embj.2021107821",
"article-title": "TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells",
"author": "Koch",
"doi-asserted-by": "crossref",
"issue": "16",
"journal-title": "Embo j",
"key": "10.1016/j.mcpro.2025.101314_bib27",
"volume": "40",
"year": "2021"
},
{
"DOI": "10.1080/22221751.2022.2117098",
"article-title": "Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "2275",
"issue": "1",
"journal-title": "Emerg Microbes Infect",
"key": "10.1016/j.mcpro.2025.101314_bib28",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.3389/fcimb.2020.589505",
"article-title": "Cathepsin L in COVID-19: from pharmacological evidences to genetics",
"author": "Gomes",
"doi-asserted-by": "crossref",
"journal-title": "Frontiers in cellular and infection microbiology",
"key": "10.1016/j.mcpro.2025.101314_bib29",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.pharmthera.2020.107587",
"article-title": "Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients",
"author": "Liu",
"doi-asserted-by": "crossref",
"journal-title": "Pharmacology & therapeutics",
"key": "10.1016/j.mcpro.2025.101314_bib30",
"volume": "213",
"year": "2020"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "269",
"issue": "3",
"journal-title": "Cell Res",
"key": "10.1016/j.mcpro.2025.101314_bib31",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1038/s42003-022-03841-8",
"article-title": "Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture",
"author": "Yuan",
"doi-asserted-by": "crossref",
"first-page": "958",
"issue": "1",
"journal-title": "Commun Biol",
"key": "10.1016/j.mcpro.2025.101314_bib32",
"volume": "5",
"year": "2022"
},
{
"DOI": "10.1371/journal.ppat.1009212",
"article-title": "Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2",
"author": "Ou",
"doi-asserted-by": "crossref",
"first-page": "e1009212",
"issue": "1",
"journal-title": "PLoS pathogens",
"key": "10.1016/j.mcpro.2025.101314_bib33",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1038/nprot.2014.138",
"article-title": "The cellular thermal shift assay for evaluating drug target interactions in cells",
"author": "Jafari",
"doi-asserted-by": "crossref",
"first-page": "2100",
"issue": "9",
"journal-title": "Nature protocols",
"key": "10.1016/j.mcpro.2025.101314_bib34",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1016/j.jsb.2010.09.007",
"article-title": "Structural basis for reversible and irreversible inhibition of human cathepsin L by their respective dipeptidyl glyoxal and diazomethylketone inhibitors",
"author": "Shenoy",
"doi-asserted-by": "crossref",
"first-page": "14",
"issue": "1",
"journal-title": "Journal of structural biology",
"key": "10.1016/j.mcpro.2025.101314_bib35",
"volume": "173",
"year": "2011"
},
{
"DOI": "10.3390/molecules25030698",
"article-title": "A review of small molecule inhibitors and functional probes of human cathepsin L",
"author": "Dana",
"doi-asserted-by": "crossref",
"first-page": "698",
"issue": "3",
"journal-title": "Molecules",
"key": "10.1016/j.mcpro.2025.101314_bib36",
"volume": "25",
"year": "2020"
},
{
"DOI": "10.1016/0006-2952(74)90174-9",
"article-title": "Lysosomotropic agents",
"author": "De Duve",
"doi-asserted-by": "crossref",
"first-page": "2495",
"issue": "18",
"journal-title": "Biochemical pharmacology",
"key": "10.1016/j.mcpro.2025.101314_bib37",
"volume": "23",
"year": "1974"
},
{
"DOI": "10.1073/pnas.2117576119",
"article-title": "SNX27 suppresses SARS-CoV-2 infection by inhibiting viral lysosome/late endosome entry",
"author": "Yang",
"doi-asserted-by": "crossref",
"issue": "4",
"journal-title": "Proceedings of the National Academy of Sciences",
"key": "10.1016/j.mcpro.2025.101314_bib38",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1128/JVI.00415-08",
"article-title": "Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide",
"author": "Bosch",
"doi-asserted-by": "crossref",
"first-page": "8887",
"issue": "17",
"journal-title": "J Virol",
"key": "10.1016/j.mcpro.2025.101314_bib39",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1074/jbc.M508381200",
"article-title": "SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "3198",
"issue": "6",
"journal-title": "J Biol Chem",
"key": "10.1016/j.mcpro.2025.101314_bib40",
"volume": "281",
"year": "2006"
},
{
"DOI": "10.1073/pnas.0505577102",
"article-title": "Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry",
"author": "Simmons",
"doi-asserted-by": "crossref",
"first-page": "11876",
"issue": "33",
"journal-title": "Proc Natl Acad Sci U S A",
"key": "10.1016/j.mcpro.2025.101314_bib41",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1016/S2213-2600(21)00222-8",
"article-title": "Colchicine for community-treated patients with COVID-19 (COLCORONA): a phase 3, randomised, double-blinded, adaptive, placebo-controlled, multicentre trial",
"author": "Tardif",
"doi-asserted-by": "crossref",
"first-page": "924",
"issue": "8",
"journal-title": "The Lancet Respiratory Medicine",
"key": "10.1016/j.mcpro.2025.101314_bib42",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1111/jep.13614",
"article-title": "Chinese herbal medicine (“3 medicines and 3 formulations”) for COVID‐19: rapid systematic review and meta‐analysis",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "13",
"issue": "1",
"journal-title": "Journal of evaluation in clinical practice",
"key": "10.1016/j.mcpro.2025.101314_bib43",
"volume": "28",
"year": "2022"
}
],
"reference-count": 43,
"references-count": 43,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S153594762500413X"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy"
}