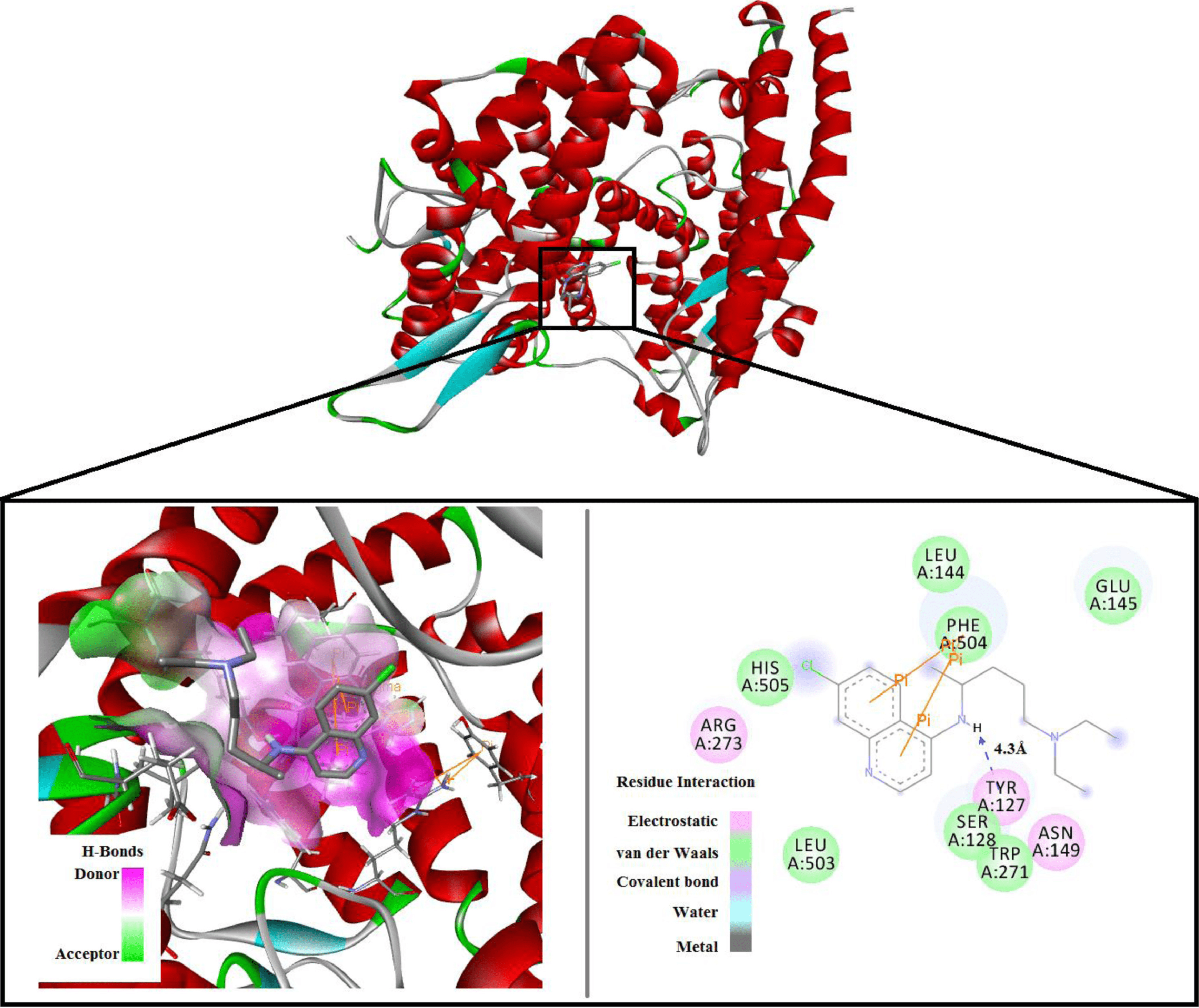
Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies
et al., Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891, Jan 2021
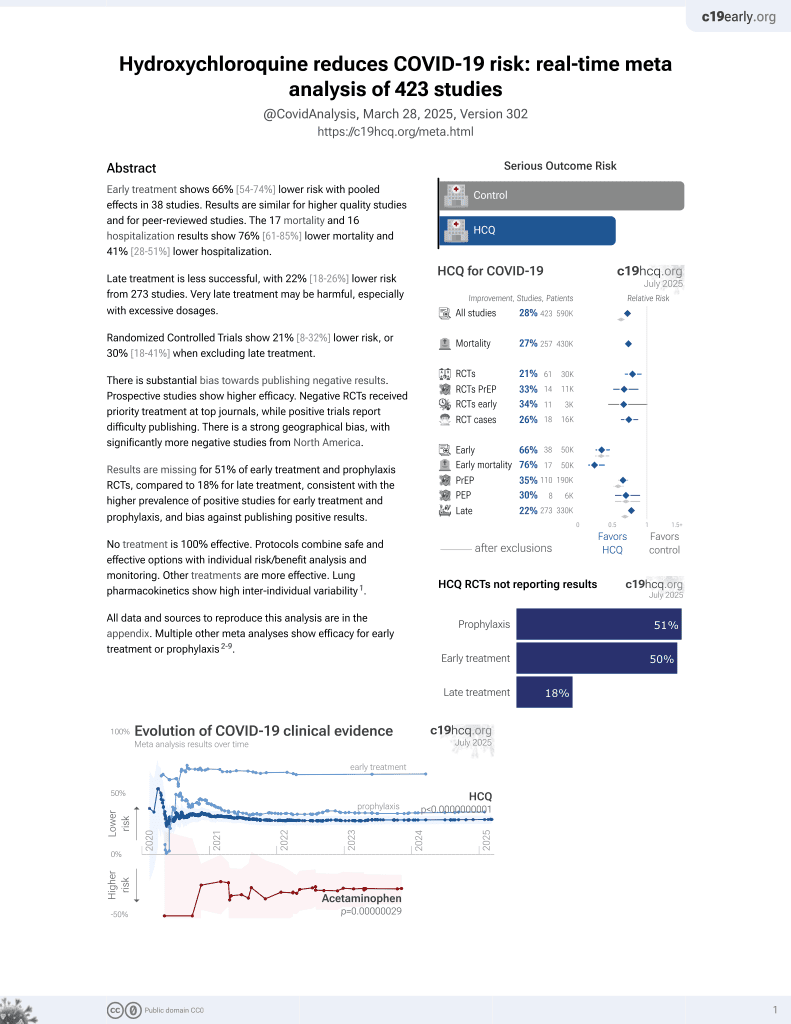
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 424 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Molecular docking study of 16 drugs showing CQ had the highest binding affinity with ACE2, and molecular dynamics study of the docked CQ-ACE2 structure. Authors conclude that CQ binds reasonably strongly with ACE2 and the stable ACE2-CQ may prevent further binding of ACE2 with the SARS-CoV-2 spike protein.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Baildya et al., 7 Jan 2021, peer-reviewed, 3 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Inhibitory capacity of chloroquine against SARS-COV-2 by effective binding with angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies
Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891
The main binding site for SARS-COV-2 spike protein in human body is human Angiotensin converting enzyme 2 (ACE2) protein receptor. Herein we present the effect of chloroquine (CLQ) on human ACE2 receptor. Molecular docking studies showed that chloroquine have a docking score is quite high compare to other well known drugs. Furthermore, molecular dynamics (MD) studies with CLQ docked ACE2 results in large fluctuations on RMSD up to 2.3 ns, indicating conformational and rotational changes due to the presence of drug molecule in the ACE2 moiety. Analysis of results showed that CLQ can effect the conformation of human ACE2 receptor. We believed that this work will help researchers to understand better the effect of CLQ on ACE2.
References
Abraham, Gready, Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5, J Comput Chem
Baildya, Ghosh, Chattopadhyay, Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies, J Mol Struct
Berendsen, Van Der Spoel, Van Drunen, GROMACS: a message-passing parallel molecular dynamics implementation, Comput Phys Commun
Biot, Daher, Chavain, Fandeur, Khalife et al., Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities, J. Med. Chem
Boonstra, Onck, Van Der Giessen, CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state, The journal of physical chemistry B
Bosseboeuf, Aubry, Nhan, Pina, Rolain et al., Azithromycin inhibits the replication of Zika virus, J Antivirals Antiretrovirals
Delvecchio, Higa, Pezzuto, Valadão, Garcez et al., Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models, Viruses
Devaux, Rolain, Raoult, ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome, Journal of Microbiology, Immunology and Infection
Fantini, Di Scala, Chahinian, Yahi, Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection, Int. J. Antimicrob. Agents
Felsenstein, Herbert, Mcnamara, Hedrich, COVID-19: immunology and treatment options, Clinical Immunology
Furuta, Komeno, Nakamura, Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase, Proceedings of the Japan Academy, Series B
Gao, Tian, Yangbreakthrough, Chloroquine Phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends, doi:10.5582/bst.2020.01047
Gautret, Lagier, Parola, Meddeb, Mailhe et al., Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int. J. Antimicrob. Agents
Huang, Wang, Li, Ren, Zhao et al., Clinical features of patients infected with 2019 novel coronavirus in, The lancet
Kandeel, Al-Nazawi, Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease, Life Sci
Lee, Tran, Allsopp, Lim, Hénin et al., CHARMM36 united atom chain model for lipids and surfactants, The journal of physical chemistry B
Liu, Cao, Xu, Wang, Zhang et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discov
Liu, Zhou, Li, Garner, Watkins et al., Research and Development On Therapeutic Agents and Vaccines For COVID-19 and Related Human Coronavirus Diseases
Marmor, Kellner, Lai, Melles, Mieler, Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision), Ophthalmology
Mcbride, Van Zyl, Fielding, The coronavirus nucleocapsid is a multifunctional protein, Viruses
Peiris, Lai, Poon, Guan, Yam et al., Coronavirus as a possible cause of severe acute respiratory syndrome, The Lancet
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera-A visualization system for exploratory research and analysis, J Comput Chem
Retallack, Di Lullo, Arias, Knopp, Laurie et al., Zika virus cell tropism in the developing human brain and inhibition by azithromycin, Proceedings of the National Academy of Sciences
Savarino, Boelaert, Cassone, Majori, Cauda, Effects of chloroquine on viral infections: an old drug against today's diseases, The Lancet infectious diseases
Stadler, Masignani, Eickmann, Becker, Abrignani et al., SARS-Beginning to understand a new virus, Nature Reviews Microbiology
Tahir, Shah, Zaman, Khan, A Dynamic Compartmental Mathematical Model Describing The Transmissibility Of MERS-CoV Virus In Public, Punjab University Journal of Mathematics
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem
Velthuis, Van Den Worm, Sims, Baric, Snijder et al., Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture, PLoS Pathog
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res
Wang, Zhang, Du, Du, Zhao et al., Remdesivir in Adults With Severe COVID-19: a randomised, Double-Blind, placebo-controlled, multicentre trial, The Lancet
Xue, Moyer, Peng, Wu, Hannafon et al., Chloroquine is a zinc ionophore, PLoS ONE
Zumla, Hui, Perlman, Middle East respiratory syndrome, The Lancet
DOI record:
{
"DOI": "10.1016/j.molstruc.2021.129891",
"ISSN": [
"0022-2860"
],
"URL": "http://dx.doi.org/10.1016/j.molstruc.2021.129891",
"alternative-id": [
"S0022286021000223"
],
"article-number": "129891",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Inhibitory capacity of chloroquine against SARS-COV-2 by effective binding with angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Journal of Molecular Structure"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.molstruc.2021.129891"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2021 Elsevier B.V. All rights reserved."
}
],
"author": [
{
"affiliation": [],
"family": "Baildya",
"given": "Nabajyoti",
"sequence": "first"
},
{
"affiliation": [],
"family": "Ghosh",
"given": "Narendra Nath",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chattopadhyay",
"given": "Asoke P.",
"sequence": "additional"
}
],
"container-title": "Journal of Molecular Structure",
"container-title-short": "Journal of Molecular Structure",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2021,
1,
8
]
],
"date-time": "2021-01-08T17:22:13Z",
"timestamp": 1610126533000
},
"deposited": {
"date-parts": [
[
2023,
10,
17
]
],
"date-time": "2023-10-17T07:02:52Z",
"timestamp": 1697526172000
},
"indexed": {
"date-parts": [
[
2024,
3,
11
]
],
"date-time": "2024-03-11T23:56:35Z",
"timestamp": 1710201395833
},
"is-referenced-by-count": 17,
"issued": {
"date-parts": [
[
2021,
4
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
},
{
"URL": "https://doi.org/10.15223/policy-017",
"content-version": "stm-asf",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
},
{
"URL": "https://doi.org/10.15223/policy-037",
"content-version": "stm-asf",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
},
{
"URL": "https://doi.org/10.15223/policy-012",
"content-version": "stm-asf",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
},
{
"URL": "https://doi.org/10.15223/policy-029",
"content-version": "stm-asf",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
},
{
"URL": "https://doi.org/10.15223/policy-004",
"content-version": "stm-asf",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
4,
1
]
],
"date-time": "2021-04-01T00:00:00Z",
"timestamp": 1617235200000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0022286021000223?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0022286021000223?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "129891",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2021,
4
]
]
},
"published-print": {
"date-parts": [
[
2021,
4
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"author": "Liu",
"key": "10.1016/j.molstruc.2021.129891_bib0001",
"series-title": "Research and Development On Therapeutic Agents and Vaccines For COVID-19 and Related Human Coronavirus Diseases",
"year": "2020"
},
{
"DOI": "10.1038/nrmicro775",
"article-title": "SARS—Beginning to understand a new virus",
"author": "Stadler",
"doi-asserted-by": "crossref",
"first-page": "209",
"issue": "3",
"journal-title": "Nature Reviews Microbiology",
"key": "10.1016/j.molstruc.2021.129891_bib0002",
"volume": "1",
"year": "2003"
},
{
"DOI": "10.1016/S0140-6736(03)13077-2",
"article-title": "Coronavirus as a possible cause of severe acute respiratory syndrome",
"author": "Peiris",
"doi-asserted-by": "crossref",
"first-page": "1319",
"issue": "9366",
"journal-title": "The Lancet",
"key": "10.1016/j.molstruc.2021.129891_bib0003",
"volume": "361",
"year": "2003"
},
{
"article-title": "A Dynamic Compartmental Mathematical Model Describing The Transmissibility Of MERS-CoV Virus In Public",
"author": "Tahir",
"first-page": "57",
"issue": "4",
"journal-title": "Punjab University Journal of Mathematics",
"key": "10.1016/j.molstruc.2021.129891_bib0004",
"volume": "51",
"year": "2019"
},
{
"DOI": "10.1016/S0140-6736(15)60454-8",
"article-title": "Middle East respiratory syndrome",
"author": "Zumla",
"doi-asserted-by": "crossref",
"first-page": "995",
"issue": "9997",
"journal-title": "The Lancet",
"key": "10.1016/j.molstruc.2021.129891_bib0005",
"volume": "386",
"year": "2015"
},
{
"DOI": "10.3390/v6082991",
"article-title": "The coronavirus nucleocapsid is a multifunctional protein",
"author": "McBride",
"doi-asserted-by": "crossref",
"first-page": "2991",
"issue": "8",
"journal-title": "Viruses",
"key": "10.1016/j.molstruc.2021.129891_bib0006",
"volume": "6",
"year": "2014"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105949",
"article-title": "Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial",
"author": "Gautret",
"doi-asserted-by": "crossref",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "10.1016/j.molstruc.2021.129891_bib0007",
"year": "2020"
},
{
"author": "Wang",
"key": "10.1016/j.molstruc.2021.129891_bib0008",
"series-title": "Remdesivir in Adults With Severe COVID-19: a randomised, Double-Blind",
"year": "2020"
},
{
"DOI": "10.2183/pjab.93.027",
"article-title": "Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase",
"author": "Furuta",
"doi-asserted-by": "crossref",
"first-page": "449",
"issue": "7",
"journal-title": "Proceedings of the Japan Academy, Series B",
"key": "10.1016/j.molstruc.2021.129891_bib0009",
"volume": "93",
"year": "2017"
},
{
"DOI": "10.1073/pnas.1618029113",
"article-title": "Zika virus cell tropism in the developing human brain and inhibition by azithromycin",
"author": "Retallack",
"doi-asserted-by": "crossref",
"first-page": "14408",
"issue": "50",
"journal-title": "Proceedings of the National Academy of Sciences",
"key": "10.1016/j.molstruc.2021.129891_bib0010",
"volume": "113",
"year": "2016"
},
{
"DOI": "10.4172/1948-5964.1000173",
"article-title": "Azithromycin inhibits the replication of Zika virus",
"author": "Bosseboeuf",
"doi-asserted-by": "crossref",
"first-page": "6",
"issue": "1",
"journal-title": "J Antivirals Antiretrovirals",
"key": "10.1016/j.molstruc.2021.129891_bib0011",
"volume": "10",
"year": "2018"
},
{
"DOI": "10.1016/j.lfs.2020.117627",
"article-title": "Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease",
"author": "Kandeel",
"doi-asserted-by": "crossref",
"journal-title": "Life Sci.",
"key": "10.1016/j.molstruc.2021.129891_bib0012",
"year": "2020"
},
{
"DOI": "10.1038/s41421-020-0156-0",
"article-title": "Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "16",
"issue": "1",
"journal-title": "Cell Discov",
"key": "10.1016/j.molstruc.2021.129891_bib0013",
"volume": "6",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"article-title": "Clinical features of patients infected with 2019 novel coronavirus in",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "497",
"issue": "10223",
"journal-title": "The lancet",
"key": "10.1016/j.molstruc.2021.129891_bib0014",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.5582/bst.2020.01047",
"doi-asserted-by": "crossref",
"key": "10.1016/j.molstruc.2021.129891_bib0015",
"unstructured": "Gao J., Tian Z., Yangbreakthrough X., Chloroquine Phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends 14 (1) (2020) 72–73, doi:10.5582/bst.2020.01047. Epub 2020 Feb 19. PMID: 32074550."
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "269",
"issue": "3",
"journal-title": "Cell Res.",
"key": "10.1016/j.molstruc.2021.129891_bib0016",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1021/jm0601856",
"article-title": "Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities",
"author": "Biot",
"doi-asserted-by": "crossref",
"first-page": "2845",
"issue": "9",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.molstruc.2021.129891_bib0017",
"volume": "49",
"year": "2006"
},
{
"DOI": "10.1016/j.ophtha.2016.01.058",
"article-title": "Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision)",
"author": "Marmor",
"doi-asserted-by": "crossref",
"first-page": "1386",
"issue": "6",
"journal-title": "Ophthalmology",
"key": "10.1016/j.molstruc.2021.129891_bib0018",
"volume": "123",
"year": "2016"
},
{
"DOI": "10.1002/jcc.20084",
"article-title": "UCSF Chimera—A visualization system for exploratory research and analysis",
"author": "Pettersen",
"doi-asserted-by": "crossref",
"first-page": "1605",
"issue": "13",
"journal-title": "J Comput Chem",
"key": "10.1016/j.molstruc.2021.129891_bib0019",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "crossref",
"first-page": "455",
"issue": "2",
"journal-title": "J Comput Chem",
"key": "10.1016/j.molstruc.2021.129891_bib0020",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1016/0010-4655(95)00042-E",
"article-title": "GROMACS: a message-passing parallel molecular dynamics implementation",
"author": "Berendsen",
"doi-asserted-by": "crossref",
"first-page": "43",
"issue": "1–3",
"journal-title": "Comput Phys Commun",
"key": "10.1016/j.molstruc.2021.129891_bib0021",
"volume": "91",
"year": "1995"
},
{
"DOI": "10.1021/jp410344g",
"article-title": "CHARMM36 united atom chain model for lipids and surfactants",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "547",
"issue": "2",
"journal-title": "The journal of physical chemistry B",
"key": "10.1016/j.molstruc.2021.129891_bib0022",
"volume": "118",
"year": "2014"
},
{
"DOI": "10.1021/acs.jpcb.6b01316",
"article-title": "CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state",
"author": "Boonstra",
"doi-asserted-by": "crossref",
"first-page": "3692",
"issue": "15",
"journal-title": "The journal of physical chemistry B",
"key": "10.1016/j.molstruc.2021.129891_bib0023",
"volume": "120",
"year": "2016"
},
{
"DOI": "10.1002/jcc.21773",
"article-title": "Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5",
"author": "Abraham",
"doi-asserted-by": "crossref",
"first-page": "2031",
"issue": "9",
"journal-title": "J Comput Chem",
"key": "10.1016/j.molstruc.2021.129891_bib0024",
"volume": "32",
"year": "2011"
},
{
"DOI": "10.1016/j.molstruc.2020.128595",
"article-title": "Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies",
"author": "Baildya",
"doi-asserted-by": "crossref",
"journal-title": "J Mol Struct",
"key": "10.1016/j.molstruc.2021.129891_bib0025",
"year": "2020"
},
{
"DOI": "10.3390/v8120322",
"article-title": "Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models",
"author": "Delvecchio",
"doi-asserted-by": "crossref",
"first-page": "322",
"issue": "12",
"journal-title": "Viruses",
"key": "10.1016/j.molstruc.2021.129891_bib0026",
"volume": "8",
"year": "2016"
},
{
"DOI": "10.1371/journal.pone.0109180",
"article-title": "Chloroquine is a zinc ionophore",
"author": "Xue",
"doi-asserted-by": "crossref",
"issue": "10",
"journal-title": "PLoS ONE",
"key": "10.1016/j.molstruc.2021.129891_bib0027",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105960",
"article-title": "Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection",
"author": "Fantini",
"doi-asserted-by": "crossref",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "10.1016/j.molstruc.2021.129891_bib0028",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1001176",
"article-title": "Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture",
"author": "Te Velthuis",
"doi-asserted-by": "crossref",
"issue": "11",
"journal-title": "PLoS Pathog.",
"key": "10.1016/j.molstruc.2021.129891_bib0029",
"volume": "6",
"year": "2010"
},
{
"DOI": "10.1016/j.jmii.2020.04.015",
"article-title": "ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome",
"author": "Devaux",
"doi-asserted-by": "crossref",
"journal-title": "Journal of Microbiology, Immunology and Infection",
"key": "10.1016/j.molstruc.2021.129891_bib0030",
"year": "2020"
},
{
"DOI": "10.1016/S1473-3099(03)00806-5",
"article-title": "Effects of chloroquine on viral infections: an old drug against today's diseases",
"author": "Savarino",
"doi-asserted-by": "crossref",
"first-page": "722",
"issue": "11",
"journal-title": "The Lancet infectious diseases",
"key": "10.1016/j.molstruc.2021.129891_bib0031",
"volume": "3",
"year": "2003"
},
{
"DOI": "10.1016/j.clim.2020.108448",
"article-title": "COVID-19: immunology and treatment options",
"author": "Felsenstein",
"doi-asserted-by": "crossref",
"journal-title": "Clinical Immunology",
"key": "10.1016/j.molstruc.2021.129891_bib0032",
"year": "2020"
}
],
"reference-count": 32,
"references-count": 32,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0022286021000223"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Inhibitory capacity of chloroquine against SARS-COV-2 by effective binding with angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1016/elsevier_cm_policy",
"volume": "1230"
}