Viral and Host Transcriptomes in SARS-CoV-2-Infected Human Lung Cells
et al., Journal of Virology, doi:10.1128/jvi.00600-21, Aug 2021
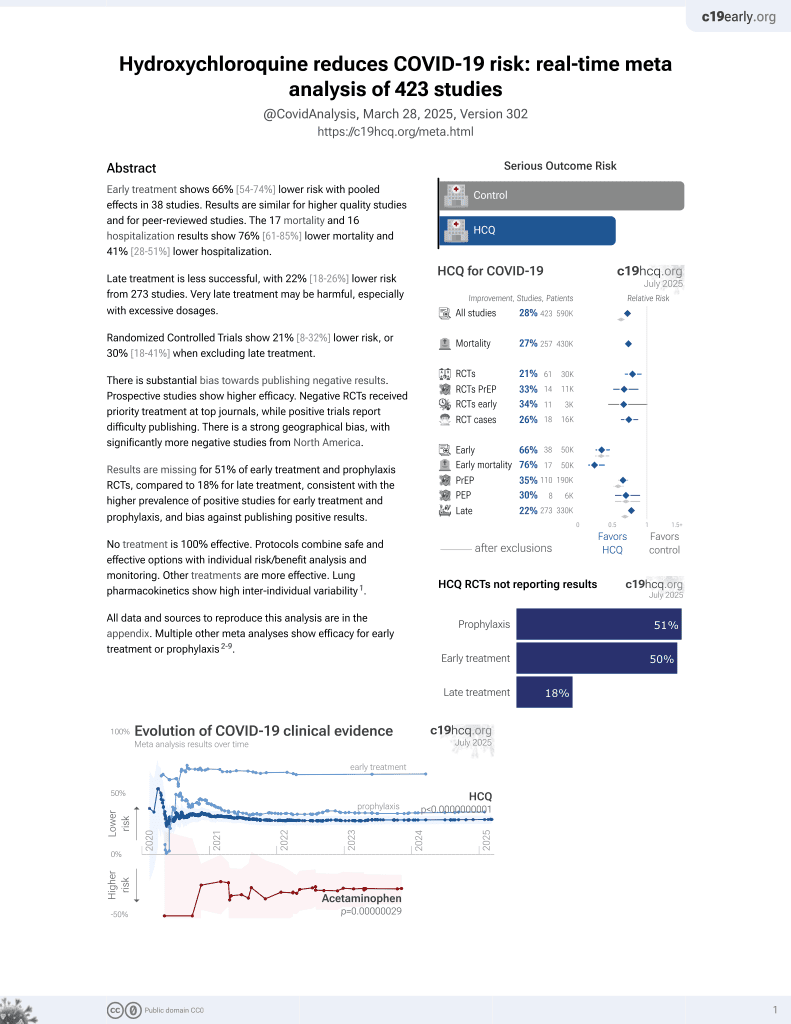
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 424 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
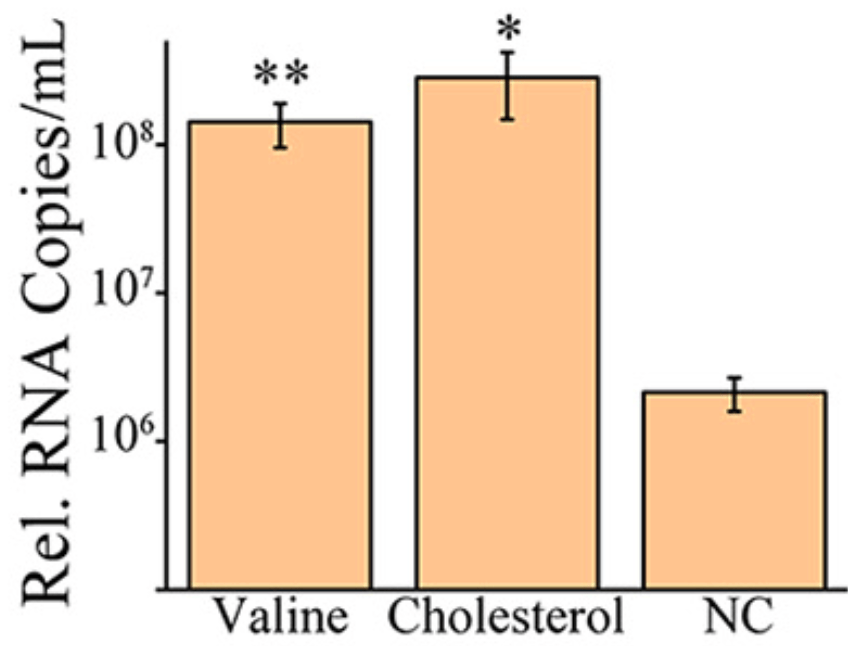
In vitro study showing that additional treatment with high levels of cholesterol enhanced viral replication, and that inhibition of host cell cholesterol metabolism promotes SARS-CoV-2 infection. Using RNA-sequencing, authors find SARS-CoV-2 infection downregulates cholesterol synthesis pathways in human lung cells. The study also highlights valine metabolism, TNF signaling and early cytokine responses, but the links between those pathways and infection severity were less clear. Overall, the data suggest host cholesterol metabolism influences SARS-CoV-2 infection, and its modulation could affect COVID-19 severity.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Wang et al., 25 Aug 2021, peer-reviewed, 12 authors.
Contact: yuwei0901@outlook.com.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Viral and Host Transcriptomes in SARS-CoV-2-Infected Human Lung Cells
doi:10.1128/JVI
Coronaviruses are commonly characterized by a unique discontinuous RNA transcriptional synthesis strategy guided by transcription-regulating sequences (TRSs). However, the details of RNA synthesis in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have not been fully elucidated. Here, we present a timescaled, gene-comparable transcriptome of SARS-CoV-2, demonstrating that ACGAAC functions as a core TRS guiding the discontinuous RNA synthesis of SARS-CoV-2 from a holistic perspective. During infection, viral transcription, rather than genome replication, dominates all viral RNA synthesis activities. The most highly expressed viral gene is the nucleocapsid gene, followed by ORF7 and ORF3 genes, while the envelope gene shows the lowest expression. Host transcription dysregulation keeps exacerbating after viral RNA synthesis reaches a maximum. The most enriched host pathways are metabolism related. Two of them (cholesterol and valine metabolism) affect viral replication in reverse. Furthermore, the activation of numerous cytokines emerges before large-scale viral RNA synthesis. IMPORTANCE SARS-CoV-2 is responsible for the current severe global health emergency that began at the end of 2019. Although the universal transcriptional strategies of coronaviruses are preliminarily understood, the details of RNA synthesis, especially the timematched transcription level of each SARS-CoV-2 gene and the principles of subgenomic mRNA synthesis, are not clear. The coterminal subgenomic mRNAs of SARS-CoV-2 present obstacles in identifying the expression of most genes by PCR-based methods, which are exacerbated by the lack of related antibodies. Moreover, SARS-CoV-2-related metabolic imbalance and cytokine storm are receiving increasing attention from both clinical and mechanistic perspectives. Our transcriptomic research provides information on both viral RNA synthesis and host responses, in which the transcription-regulating sequences and transcription levels of viral genes are demonstrated, and the metabolic dysregulation and cytokine levels identified at the host cellular level support the development of novel medical treatment strategies.
were selected and referred to as short "query reads" (see Fig. S1 ). Their first nt were located one by one downstream at a specific region of the 59 UTR. By querying the combined read pool containing all viral sequences, all sequences of 30 nt in length whose 59 15-nt sequences were identical to the query reads were returned and their numbers were counted. The 15-nt sequences downstream of the corresponding query reads were referred to as their "return reads." Located around the possible TRS, the return reads could be either manually aligned continuously to gRNAs or aligned discontinuously to sgmRNAs with the 59 partial sequence homologous to the leader UTR (upstream of the leader TRS) and the 39 sequence homologous to various ORFs (downstream of the body TRS). When a site could be regarded as either 59 continuous (continuous to upstream query reads) or 39 continuous (continuous to downstream ORFs), it was designated 59 continuous, as we intended to identify the probable leader TRS as long as possible. In parallel, two additional types of 15-nt query reads with sequences homologous to the beginning of the ORFs were used: the first type started at the body TRS (6 nt) and ended at 19 nt of the downstream ORF (referred to as "in-TRS reads"), and the second type was homologous to 11 to 115 nt of the ORFs adjacent to the downstream body TRS (referred to as "after-TRS reads"). For each known ORF, an in-TRS read and an after-TRS read were..
References
Baker, Reddy, Modulation of life and death by the TNF receptor superfamily, Oncogene, doi:10.1038/sj.onc.1202568
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell, doi:10.1016/j.cell.2020.04.026
Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature, doi:10.1038/s41586-020-2332-7
Chang, Guarente, SIRT1 and other sirtuins in metabolism, Trends Endocrinol Metab, doi:10.1016/j.tem.2013.12.001
Chua, Lukassen, Trump, Hennig, Wendisch et al., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis, Nat Biotechnol, doi:10.1038/s41587-020-0602-4
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell, doi:10.1016/j.cell.2020.10.030
Hachim, Kavian, Cohen, Chin, Chu et al., ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection, Nat Immunol, doi:10.1038/s41590-020-0773-7
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Kim, Lee, Yang, Kim, Kim et al., The Architecture of SARS-CoV-2 transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Lee, Cao, Mostoslavsky, Lombard, Liu et al., A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy, Proc Natl Acad Sci U S A, doi:10.1073/pnas.0712145105
Lee, Park, Jeong, Ahn, Choi et al., Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19, Sci Immunol, doi:10.1126/sciimmunol.abd1554
Li, Zhang, Blander, Tse, Krieger et al., SIRT1 deacetylates and positively regulates the nuclear receptor LXR, Mol Cell, doi:10.1016/j.molcel.2007.07.032
Liao, Liu, Yuan, Wen, Xu et al., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat Med, doi:10.1038/s41591-020-0901-9
Livak, Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 22DDCT method, Methods, doi:10.1006/meth.2001.1262
Medler, Wajant, Tumor necrosis factor receptor-2 (TNFR2): an overview of an emerging drug target, Expert Opin Ther Targets, doi:10.1080/14728222.2019.1586886
Miyazaki, Ichiki, Hashimoto, Inanaga, Imayama et al., SIRT1, a longevity gene, downregulates angiotensin II type 1 receptor expression in vascular smooth muscle cells, Arterioscler Thromb Vasc Biol, doi:10.1161/ATVBAHA.108.166991
Neinast, Murashige, Arany, Branched chain amino acids, Annu Rev Physiol, doi:10.1146/annurev-physiol-020518-114455
Radenkovic, Chawla, Pirro, Sahebkar, Banach, Cholesterol in relation to COVID-19: should we care about it?, J Clin Med, doi:10.3390/jcm9061909
Salomon, Hoffmann, Webster, Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection, Proc Natl Acad Sci U S A, doi:10.1073/pnas.0705289104
Snijder, Bredenbeek, Dobbe, Thiel, Ziebuhr et al., Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage, J Mol Biol, doi:10.1016/S0022-2836(03)00865-9
Snijder, Decroly, Ziebuhr, Chapter three -the nonstructural proteins directing coronavirus RNA synthesis and processing
Sola, Almazán, Zúñiga, Enjuanes, Continuous and discontinuous RNA synthesis in coronaviruses, Annu Rev Virol, doi:10.1146/annurev-virology-100114-055218
Storz, Forkhead homeobox type O transcription factors in the responses to oxidative stress, Antioxid Redox Signal, doi:10.1089/ars.2010.3405
Stukalov, Girault, Grass, Bergant, Karayel et al., Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV, doi:10.1101/2020.06.17.156455:2020.06.17.156455
Taiaroa, Rawlinson, Featherstone, Pitt, Caly et al., Direct RNA sequencing and early evolution of SARS-CoV-2, doi:10.1101/2020.03.05.976167:2020.03.05.976167
Thiel, Ivanov, Putics, Hertzig, Schelle et al., Mechanisms and enzymes involved in SARS coronavirus genome expression, J Gen Virol, doi:10.1099/vir.0.19424-0
Wang, Hu, Hu, Zhu, Liu et al., Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China, JAMA, doi:10.1001/jama.2020.1585
Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell, doi:10.1016/j.cell.2020.12.004
Wu, Zhao, Yu, Chen, Song et al., A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Xiong, Liu, Cao, Wang, Guo et al., Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients, Emerg Microbes Infect, doi:10.1080/22221751.2020.1747363
Yao, Irwin, Zhao, Nilsen, Hamilton et al., Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease, Proc Natl Acad Sci U S A, doi:10.1073/pnas.0903563106
Zhang, Li, Cruz, Kone, Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct, J Biol Chem, doi:10.1074/jbc.M109.020073
DOI record:
{
"DOI": "10.1128/jvi.00600-21",
"ISSN": [
"0022-538X",
"1098-5514"
],
"URL": "http://dx.doi.org/10.1128/jvi.00600-21",
"abstract": "<jats:p>SARS-CoV-2 is responsible for the current severe global health emergency that began at the end of 2019. Although the universal transcriptional strategies of coronaviruses are preliminarily understood, the details of RNA synthesis, especially the time-matched transcription level of each SARS-CoV-2 gene and the principles of subgenomic mRNA synthesis, are not clear.</jats:p>",
"alternative-id": [
"10.1128/JVI.00600-21"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2021-04-07"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2021-06-02"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2021-08-25"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-3974-3421",
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"authenticated-orcid": true,
"family": "Wang",
"given": "Xuefeng",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
},
{
"name": "School of Life Sciences, Northeast Normal University, Changchun, People’s Republic of China"
}
],
"family": "Zhao",
"given": "Yudong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Yan",
"given": "Feihu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Wang",
"given": "Tiecheng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Sun",
"given": "Weiyang",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Feng",
"given": "Na",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
},
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"family": "Wang",
"given": "Wenqi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"family": "Wang",
"given": "Hongmei",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7438-0638",
"affiliation": [
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"authenticated-orcid": true,
"family": "He",
"given": "Hongbin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Yang",
"given": "Songtao",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Xia",
"given": "Xianzhu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Gao",
"given": "Yuwei",
"sequence": "additional"
}
],
"container-title": "Journal of Virology",
"container-title-short": "J Virol",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2021,
6,
13
]
],
"date-time": "2021-06-13T23:05:56Z",
"timestamp": 1623625556000
},
"deposited": {
"date-parts": [
[
2022,
3,
5
]
],
"date-time": "2022-03-05T16:28:34Z",
"timestamp": 1646497714000
},
"editor": [
{
"affiliation": [],
"family": "Subbarao",
"given": "Kanta",
"sequence": "additional"
}
],
"funder": [
{
"DOI": "10.13039/501100012166",
"award": [
"2016YFD0500203"
],
"doi-asserted-by": "publisher",
"name": "National Key Research and Development Program of China"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
21
]
],
"date-time": "2024-02-21T11:09:16Z",
"timestamp": 1708513756036
},
"is-referenced-by-count": 8,
"issue": "18",
"issued": {
"date-parts": [
[
2021,
8,
25
]
]
},
"journal-issue": {
"issue": "18",
"published-print": {
"date-parts": [
[
2021,
8,
25
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://doi.org/10.1128/ASMCopyrightv2",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
8,
25
]
],
"date-time": "2021-08-25T00:00:00Z",
"timestamp": 1629849600000
}
},
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
8,
25
]
],
"date-time": "2021-08-25T00:00:00Z",
"timestamp": 1629849600000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/JVI.00600-21",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/JVI.00600-21",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"prefix": "10.1128",
"published": {
"date-parts": [
[
2021,
8,
25
]
]
},
"published-print": {
"date-parts": [
[
2021,
8,
25
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"key": "e_1_3_3_2_2",
"unstructured": "World Health Organization. 2020. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/."
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_2"
},
{
"author": "Snijder EJ",
"first-page": "59",
"key": "e_1_3_3_4_2",
"unstructured": "Snijder EJ, Decroly E, Ziebuhr J. 2016. Chapter three - the nonstructural proteins directing coronavirus RNA synthesis and processing, p 59–126. In Ziebuhr J (ed), Advances in virus research, vol 96. Academic Press.",
"volume-title": "Advances in virus research",
"year": "2016"
},
{
"DOI": "10.1016/S0022-2836(03)00865-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1146/annurev-virology-100114-055218",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.1099/vir.0.19424-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1101/2020.03.05.976167",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_9_2",
"unstructured": "Taiaroa G Rawlinson D Featherstone L Pitt M Caly L Druce J Purcell D Harty L Tran T Roberts J Scott N Catton M Williamson D Coin L Duchene S. 2020. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv 10.1101/2020.03.05.976167:2020.03.05.976167."
},
{
"DOI": "10.1038/s41587-020-0602-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_2"
},
{
"DOI": "10.1080/22221751.2020.1747363",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_2"
},
{
"DOI": "10.1101/2020.06.17.156455",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_12_2",
"unstructured": "Stukalov A Girault V Grass V Bergant V Karayel O Urban C Haas DA Huang Y Oubraham L Wang A Hamad SM Piras A Tanzer M Hansen FM Enghleitner T Reinecke M Lavacca TM Ehmann R Wölfel R Jores J Kuster B Protzer U RR Ziebuhr J Thiel V Scaturro P Mann M Pichlmair A. 2020. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv 10.1101/2020.06.17.156455:2020.06.17.156455."
},
{
"DOI": "10.1073/pnas.0903563106",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_13_2"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"DOI": "10.1016/j.tem.2013.12.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_2"
},
{
"DOI": "10.1016/j.molcel.2007.07.032",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"DOI": "10.1074/jbc.M109.020073",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_2"
},
{
"DOI": "10.1089/ars.2010.3405",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1073/pnas.0712145105",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1001/jama.2020.1585",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1161/ATVBAHA.108.166991",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.1146/annurev-physiol-020518-114455",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1038/sj.onc.1202568",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.1038/s41591-020-0901-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.1038/s41590-020-0773-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_27_2"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.3390/jcm9061909",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_2"
},
{
"DOI": "10.1080/14728222.2019.1586886",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_2"
},
{
"DOI": "10.1126/sciimmunol.abd1554",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"DOI": "10.1073/pnas.0705289104",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_2"
},
{
"DOI": "10.1006/meth.2001.1262",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
}
],
"reference-count": 33,
"references-count": 33,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/JVI.00600-21"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Insect Science",
"Immunology",
"Microbiology"
],
"subtitle": [],
"title": "Viral and Host Transcriptomes in SARS-CoV-2-Infected Human Lung Cells",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "95"
}