Screening of an FDA-Approved Compound Library Identifies Four Small-Molecule Inhibitors of Middle East Respiratory Syndrome Coronavirus Replication in Cell Culture
et al., Antimicrobial Agents and Chemotherapy, Jul 2014, 58:8, 4875-4884, doi:10.1128/AAC.03011-14, Jul 2014
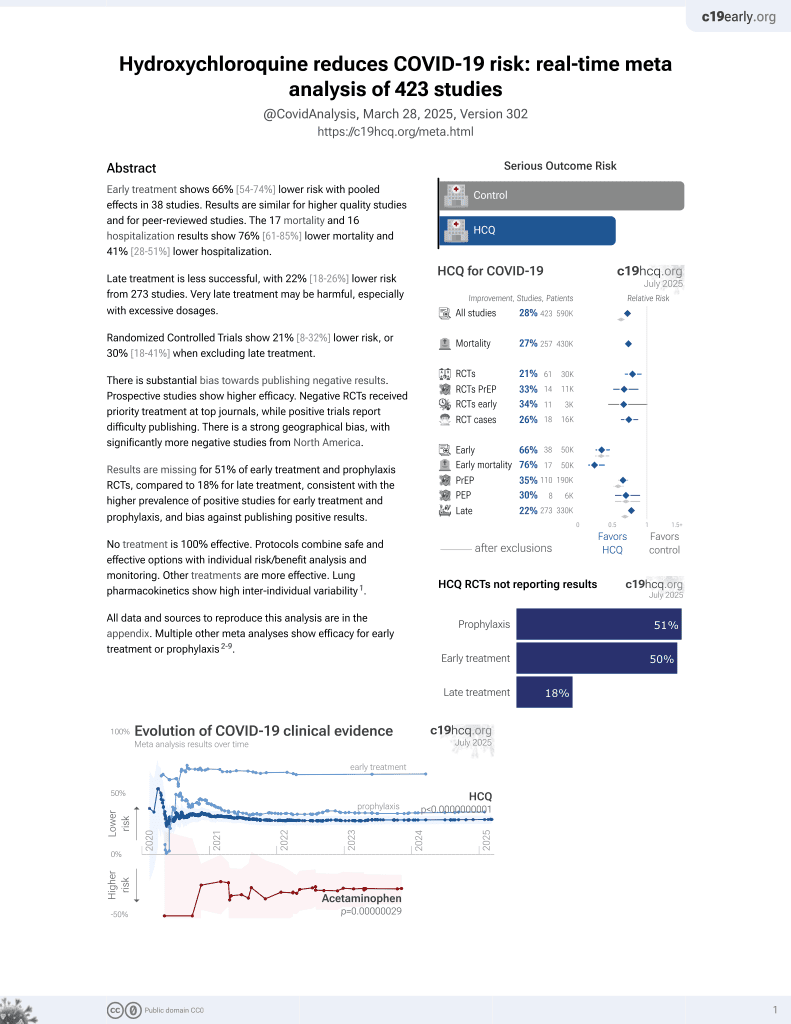
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 423 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
CQ inhibits SARS-CoV, MERS-CoV, and HCoV-229E-GFP replication in the low-micromolar range.
39 preclinical studies support the efficacy of HCQ for COVID-19:
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
de Wilde et al., 15 Jul 2014, peer-reviewed, 9 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: Screening of an FDA-Approved Compound Library Identifies Four
Small-Molecule Inhibitors of Middle East Respiratory Syndrome
Coronavirus Replication in Cell Culture
Adriaan H. de Wilde,a Dirk Jochmans,b Clara C. Posthuma,a Jessika C. Zevenhoven-Dobbe,a Stefan van Nieuwkoop,c
Theo M. Bestebroer,c Bernadette G. van den Hoogen,c Johan Neyts,b Eric J. Snijdera
Molecular Virology Laboratory, Department of Medical Microbiology, Leiden University Medical Center, Leiden, Netherlandsa; Rega Institute for Medical Research, KU,
Leuven, Belgiumb; Department of Viroscience, Erasmus Medical Center, Rotterdam, Netherlandsc
I
n June 2012, a previously unknown coronavirus was isolated
from a patient who died from acute pneumonia and renal failure
in Saudi Arabia (1, 2). Since then, the virus, now known as the
Middle East respiratory syndrome coronavirus (MERS-CoV) (3),
has been contracted by hundreds of others in geographically distinct locations in the Middle East, and evidence for limited human-to-human transmission has accumulated (4). Travel-related
MERS-CoV infections were reported from a variety of countries in
Europe, Africa, Asia, and the United States, causing small local
infection clusters in several cases (http://www.who.int/csr/disease
/coronavirus_infections/en/). About 200 laboratory-confirmed
human MERS cases were registered during the first 2 years of this
outbreak, but recently, for reasons that are poorly understood
thus far, this number has more than tripled within just 2 months’
time (April-May 2014 [5]). This sharp increase in reported infections has enhanced concerns that we might be confronted with a
repeat of the 2003 severe acute respiratory syndrome (SARS) episode, concerns aggravated by the fact that the animal reservoir for
MERS-CoV remains to be identified with certainty (6–9). Furthermore, at about 30%, the current human case fatality rate is
alarmingly high, even though many deaths were associated with
underlying medical conditions. MERS-CoV infection in humans
can cause clinical symptoms resembling SARS, such as high fever
and acute pneumonia, although the two viruses were reported to
use different entry receptors, dipeptidyl peptidase 4 (DPP4) (10)
and angiotensin-converting enzyme 2 (ACE2) (11), respectively.
Coronaviruses are currently divided across four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (12). MERS-CoV was identified as a member of lineage C
August 2014 Volume 58 Number 8
of the genus Betacoronavirus (2), which also includes coronaviruses of bat (13, 14) and hedgehog (6) origin. Following the 2003
SARS epidemic, studies into the complex genome, proteome, and
replication cycle of coronaviruses were intensified. Coronaviruses
are enveloped viruses with a positive-sense RNA genome of unprecedented length (25 to 32 kb [12, 15, 16]). The crystal structures of a substantial number of viral nonstructural and structural
proteins were solved, and targeted drug design was performed for
some of those (reviewed in reference 17). Unfortunately, thus far,
none of these efforts resulted in antiviral drugs that were advanced
beyond the preclinical phase (18). The 2003 SARS-CoV epidemic
was controlled within a few months after its onset, and since then,
the virus has not reemerged, although close relatives continue to
circulate in bat species (14). Consequently, the interest in anticoronavirus drug development has been limited, until the emer-
Received 10 April 2014 Returned for..
DOI record:
{
"DOI": "10.1128/aac.03011-14",
"ISSN": [
"0066-4804",
"1098-6596"
],
"URL": "http://dx.doi.org/10.1128/AAC.03011-14",
"abstract": "<jats:title>ABSTRACT</jats:title><jats:p>Coronaviruses can cause respiratory and enteric disease in a wide variety of human and animal hosts. The 2003 outbreak of severe acute respiratory syndrome (SARS) first demonstrated the potentially lethal consequences of zoonotic coronavirus infections in humans. In 2012, a similar previously unknown coronavirus emerged, Middle East respiratory syndrome coronavirus (MERS-CoV), thus far causing over 650 laboratory-confirmed infections, with an unexplained steep rise in the number of cases being recorded over recent months. The human MERS fatality rate of ∼30% is alarmingly high, even though many deaths were associated with underlying medical conditions. Registered therapeutics for the treatment of coronavirus infections are not available. Moreover, the pace of drug development and registration for human use is generally incompatible with strategies to combat emerging infectious diseases. Therefore, we have screened a library of 348 FDA-approved drugs for anti-MERS-CoV activity in cell culture. If such compounds proved sufficiently potent, their efficacy might be directly assessed in MERS patients. We identified four compounds (chloroquine, chlorpromazine, loperamide, and lopinavir) inhibiting MERS-CoV replication in the low-micromolar range (50% effective concentrations [EC<jats:sub>50</jats:sub>s], 3 to 8 μM). Moreover, these compounds also inhibit the replication of SARS coronavirus and human coronavirus 229E. Although their protective activity (alone or in combination) remains to be assessed in animal models, our findings may offer a starting point for treatment of patients infected with zoonotic coronaviruses like MERS-CoV. Although they may not necessarily reduce viral replication to very low levels, a moderate viral load reduction may create a window during which to mount a protective immune response.</jats:p>",
"alternative-id": [
"10.1128/AAC.03011-14"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2014-04-10"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2014-05-14"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2014-07-15"
}
],
"author": [
{
"affiliation": [
{
"name": "Molecular Virology Laboratory, Department of Medical Microbiology, Leiden University Medical Center, Leiden, Netherlands"
}
],
"family": "de Wilde",
"given": "Adriaan H.",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Rega Institute for Medical Research, KU, Leuven, Belgium"
}
],
"family": "Jochmans",
"given": "Dirk",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Molecular Virology Laboratory, Department of Medical Microbiology, Leiden University Medical Center, Leiden, Netherlands"
}
],
"family": "Posthuma",
"given": "Clara C.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Molecular Virology Laboratory, Department of Medical Microbiology, Leiden University Medical Center, Leiden, Netherlands"
}
],
"family": "Zevenhoven-Dobbe",
"given": "Jessika C.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Viroscience, Erasmus Medical Center, Rotterdam, Netherlands"
}
],
"family": "van Nieuwkoop",
"given": "Stefan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Viroscience, Erasmus Medical Center, Rotterdam, Netherlands"
}
],
"family": "Bestebroer",
"given": "Theo M.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Viroscience, Erasmus Medical Center, Rotterdam, Netherlands"
}
],
"family": "van den Hoogen",
"given": "Bernadette G.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Rega Institute for Medical Research, KU, Leuven, Belgium"
}
],
"family": "Neyts",
"given": "Johan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Molecular Virology Laboratory, Department of Medical Microbiology, Leiden University Medical Center, Leiden, Netherlands"
}
],
"family": "Snijder",
"given": "Eric J.",
"sequence": "additional"
}
],
"container-title": "Antimicrobial Agents and Chemotherapy",
"container-title-short": "Antimicrob Agents Chemother",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2014,
5,
20
]
],
"date-time": "2014-05-20T05:04:47Z",
"timestamp": 1400562287000
},
"deposited": {
"date-parts": [
[
2023,
7,
13
]
],
"date-time": "2023-07-13T07:21:24Z",
"timestamp": 1689232884000
},
"indexed": {
"date-parts": [
[
2024,
5,
9
]
],
"date-time": "2024-05-09T19:54:54Z",
"timestamp": 1715284494431
},
"is-referenced-by-count": 568,
"issue": "8",
"issued": {
"date-parts": [
[
2014,
8
]
]
},
"journal-issue": {
"issue": "8",
"published-print": {
"date-parts": [
[
2014,
8
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2014,
8,
1
]
],
"date-time": "2014-08-01T00:00:00Z",
"timestamp": 1406851200000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/AAC.03011-14",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/AAC.03011-14",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"page": "4875-4884",
"prefix": "10.1128",
"published": {
"date-parts": [
[
2014,
8
]
]
},
"published-print": {
"date-parts": [
[
2014,
8
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"DOI": "10.1056/NEJMoa1211721",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_2_2"
},
{
"DOI": "10.1128/mBio.00473-12",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_2"
},
{
"DOI": "10.1128/JVI.01244-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_4_2"
},
{
"DOI": "10.1128/mBio.01062-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1126/science.344.6183.457",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1128/JVI.01600-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.1016/S1473-3099(13)70690-X",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1016/S1473-3099(13)70164-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_9_2"
},
{
"DOI": "10.3201/eid1911.131172",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_2"
},
{
"DOI": "10.1038/nature12005",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_2"
},
{
"DOI": "10.1038/nature02145",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_12_2"
},
{
"author": "de Groot RJ",
"first-page": "785",
"key": "e_1_3_3_13_2",
"unstructured": "de GrootRJCowleyJAEnjuanesLFaabergKSPerlmanSRottierPJSnijderEJZiebuhrJGorbalenyaAE. 2012. Order of Nidovirales, p 785–795. In KingAAdamsMCarstensELefkowitzEJ (ed), Virus taxonomy, the 9th report of the International Committee on Taxonomy of Viruses. Academic Press, New York, NY.",
"volume-title": "Virus taxonomy, the 9th report of the International Committee on Taxonomy of Viruses",
"year": "2012"
},
{
"DOI": "10.1016/j.antiviral.2013.10.013",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"DOI": "10.1038/nature12711",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_2"
},
{
"DOI": "10.1128/JVI.02351-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"DOI": "10.1128/JVI.06540-11",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_2"
},
{
"DOI": "10.2174/187152609787847659",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1016/j.antiviral.2013.08.015",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.1517/13543770802609384",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1371/journal.pbio.0030324",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1021/jm1004489",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.1016/j.febslet.2004.08.015",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1016/j.febslet.2008.12.059",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.1073/pnas.0403596101",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.1073/pnas.0805240105",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"DOI": "10.1128/JVI.02105-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_27_2"
},
{
"DOI": "10.1038/nrmicro3143",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.1093/jac/dkn243",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_2"
},
{
"DOI": "10.3978/j.issn.2072-1439.2013.06.18",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_2"
},
{
"DOI": "10.1371/journal.pmed.0030343",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.1038/srep01686",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"DOI": "10.1038/nm.3362",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_2"
},
{
"DOI": "10.1016/j.ijid.2013.12.003",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
},
{
"DOI": "10.1099/vir.0.052910-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_35_2"
},
{
"DOI": "10.1128/JVI.03496-12",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_36_2"
},
{
"DOI": "10.1038/nm1001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_37_2"
},
{
"DOI": "10.1016/j.antiviral.2005.01.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_38_2"
},
{
"DOI": "10.1089/1079990041535610",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_39_2"
},
{
"DOI": "10.1128/JVI.00009-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_40_2"
},
{
"DOI": "10.1128/mBio.00611-12",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_41_2"
},
{
"DOI": "10.1016/j.jinf.2013.09.029",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_42_2"
},
{
"DOI": "10.1099/vir.0.061911-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_43_2"
},
{
"DOI": "10.1099/vir.0.034983-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_44_2"
},
{
"DOI": "10.1371/journal.ppat.1002331",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_45_2"
},
{
"DOI": "10.1186/1297-9716-43-41",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_46_2"
},
{
"DOI": "10.1073/pnas.1323279111",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_47_2"
},
{
"DOI": "10.1128/JVI.02501-05",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_48_2"
},
{
"DOI": "10.1056/NEJMoa030747",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_49_2"
},
{
"DOI": "10.1128/mBio.00171-10",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_50_2"
},
{
"DOI": "10.1371/journal.pone.0032857",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_51_2"
},
{
"DOI": "10.1016/j.bbrc.2004.08.085",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_52_2"
},
{
"DOI": "10.1186/1743-422X-2-69",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_53_2"
},
{
"DOI": "10.1016/j.antiviral.2007.10.011",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_54_2"
},
{
"DOI": "10.1128/AAC.01509-08",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_55_2"
},
{
"DOI": "10.1016/j.antiviral.2013.04.016",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_56_2"
},
{
"DOI": "10.1083/jcb.123.5.1107",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_57_2"
},
{
"DOI": "10.3201/eid1905.130057",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_58_2"
},
{
"DOI": "10.2217/fvl.11.33",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_59_2"
},
{
"DOI": "10.1016/j.imlet.2013.07.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_60_2"
},
{
"DOI": "10.1016/S1473-3099(03)00806-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_61_2"
},
{
"DOI": "10.1371/journal.pone.0060579",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_62_2"
},
{
"DOI": "10.1128/JVI.00164-09",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_63_2"
},
{
"DOI": "10.1016/0042-6822(66)90046-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_64_2"
},
{
"DOI": "10.1099/0022-1317-65-1-227",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_65_2"
},
{
"DOI": "10.1177/095632020601700505",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_66_2"
},
{
"DOI": "10.1128/AAC.02279-12",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_67_2"
},
{
"DOI": "10.1038/mp.2012.47",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_68_2"
},
{
"DOI": "10.1371/journal.pone.0028923",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_69_2"
},
{
"DOI": "10.1128/JVI.00024-06",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_70_2"
},
{
"DOI": "10.1128/JVI.00253-07",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_71_2"
},
{
"DOI": "10.1007/978-0-387-33012-9_54",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_72_2"
},
{
"DOI": "10.1128/JVI.00837-08",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_73_2"
},
{
"DOI": "10.1146/annurev.pa.24.040184.002045",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_74_2"
},
{
"DOI": "10.1016/j.jpainsymman.2011.06.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_75_2"
},
{
"DOI": "10.1146/annurev.pa.23.040183.001431",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_76_2"
},
{
"DOI": "10.1097/FTD.0b013e31822d578b",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_77_2"
},
{
"DOI": "10.2174/138161210793563446",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_78_2"
},
{
"DOI": "10.47102/annals-acadmedsg.V36N6p438",
"article-title": "Pharmacologic treatment of SARS: current knowledge and recommendations",
"author": "Tai DY",
"doi-asserted-by": "crossref",
"first-page": "438",
"journal-title": "Ann. Acad. Med. Singapore",
"key": "e_1_3_3_79_2",
"unstructured": "TaiDY. 2007. Pharmacologic treatment of SARS: current knowledge and recommendations. Ann. Acad. Med. Singapore 36:438–443.",
"volume": "36",
"year": "2007"
},
{
"DOI": "10.1016/j.antiviral.2013.08.016",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_80_2"
},
{
"DOI": "10.1056/NEJMc1215691",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_81_2"
},
{
"DOI": "10.1093/infdis/jit590",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_82_2"
},
{
"DOI": "10.1073/pnas.1310744110",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_83_2"
},
{
"DOI": "10.1099/vir.0.060640-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_84_2"
},
{
"DOI": "10.1128/JVI.02935-13",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_85_2"
},
{
"DOI": "10.1128/AAC.03036-14",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_86_2"
}
],
"reference-count": 85,
"references-count": 85,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/AAC.03011-14"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Screening of an FDA-Approved Compound Library Identifies Four Small-Molecule Inhibitors of Middle East Respiratory Syndrome Coronavirus Replication in Cell Culture",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "58"
}