Synergistic and Antagonistic Drug Combinations against SARS-CoV-2
et al., Molecular Therapy, doi:10.1016/j.ymthe.2020.12.016, Feb 2021
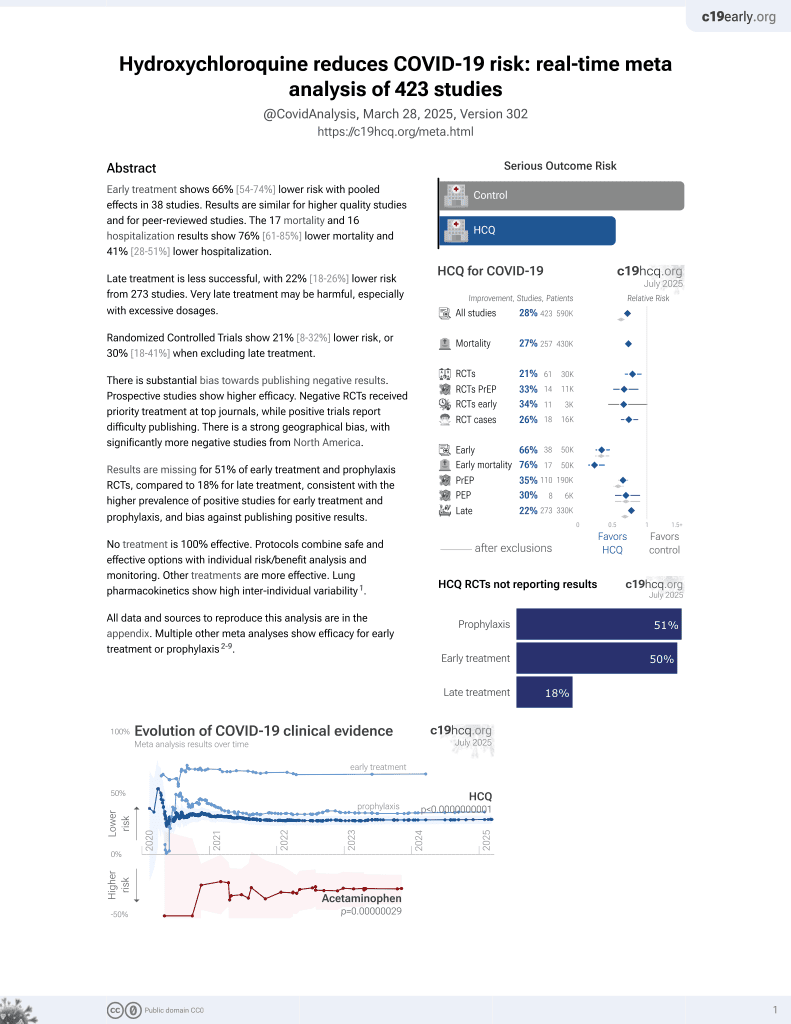
HCQ for COVID-19
1st treatment shown to reduce risk in
March 2020, now with p < 0.00000000001 from 423 studies, used in 59 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
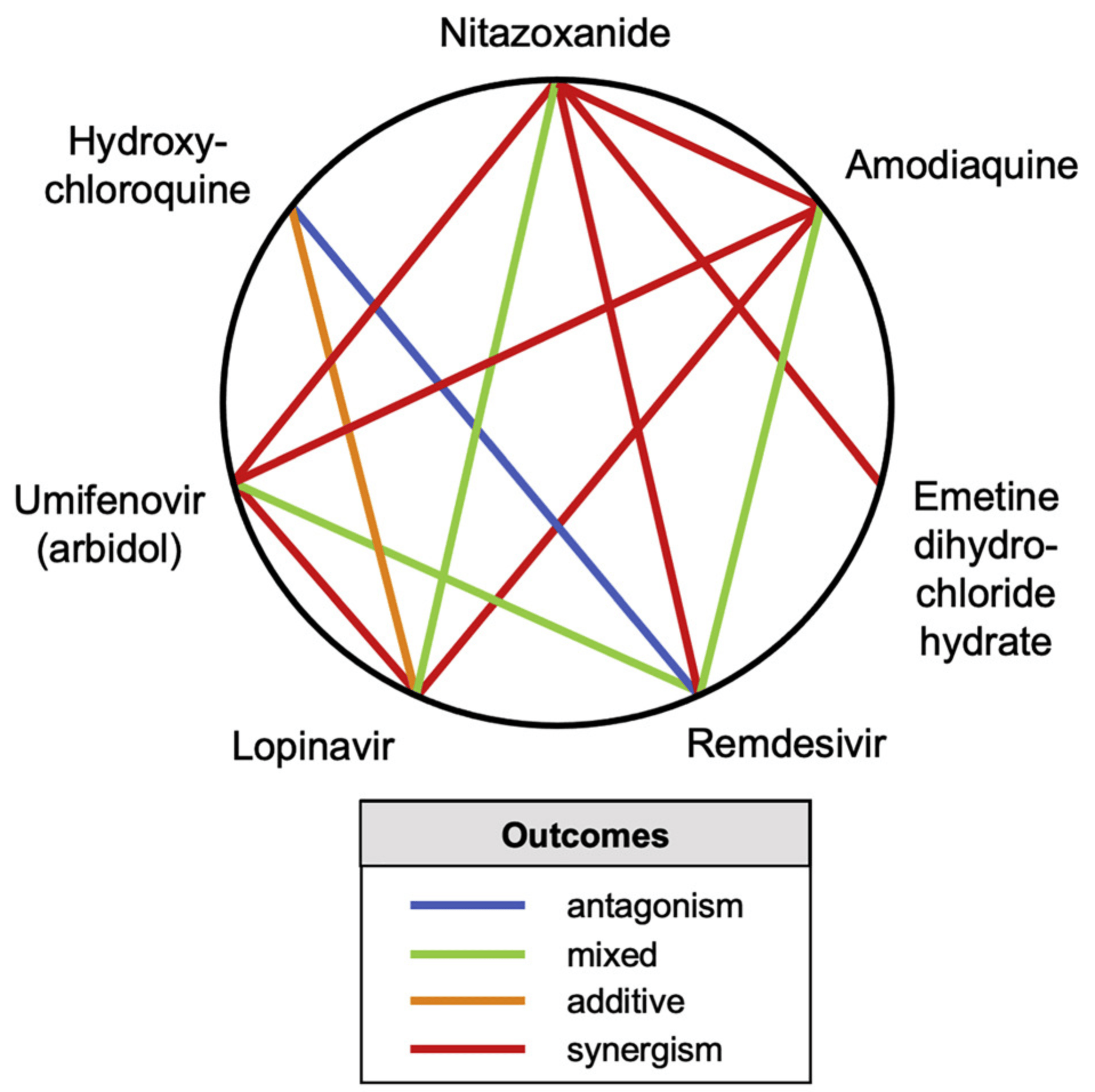
In silico and in vitro study showing both synergistic and antagonistic drug combinations against SARS-CoV-2 in Vero E6 cells. Authors screened 73 combinations of 32 drugs and identified 16 synergistic and 8 antagonistic combinations. Nitazoxanide showed promising synergy with remdesivir, amodiaquine, and umifenovir, while there was significant antagonism between remdesivir and lysosomotropic agents like hydroxychloroquine, suggesting that they should not be used together.
39 preclinical studies support the efficacy of HCQ for COVID-19:
Study covers remdesivir, nitazoxanide, and HCQ.
1.
Shang et al., Identification of Cathepsin L as the molecular target of hydroxychloroquine with chemical proteomics, Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101314.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Guimarães Silva et al., Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study, ACTA MEDICA IRANICA, doi:10.18502/acta.v61i2.12533.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
7.
Yadav et al., Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor Against SARS-CoV-2: A Computational Study, Research Square, doi:10.21203/rs.3.rs-628277/v1.
8.
Hussein et al., Molecular Docking Identification for the efficacy of Some Zinc Complexes with Chloroquine and Hydroxychloroquine against Main Protease of COVID-19, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129979.
9.
Baildya et al., Inhibitory capacity of Chloroquine against SARS-COV-2 by effective binding with Angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies, Journal of Molecular Structure, doi:10.1016/j.molstruc.2021.129891.
10.
Noureddine et al., Quantum chemical studies on molecular structure, AIM, ELF, RDG and antiviral activities of hybrid hydroxychloroquine in the treatment of COVID-19: molecular docking and DFT calculations, Journal of King Saud University - Science, doi:10.1016/j.jksus.2020.101334.
11.
Tarek et al., Pharmacokinetic Basis of the Hydroxychloroquine Response in COVID-19: Implications for Therapy and Prevention, European Journal of Drug Metabolism and Pharmacokinetics, doi:10.1007/s13318-020-00640-6.
12.
Rowland Yeo et al., Impact of Disease on Plasma and Lung Exposure of Chloroquine, Hydroxychloroquine and Azithromycin: Application of PBPK Modeling, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.1955.
13.
Hitti et al., Hydroxychloroquine attenuates double-stranded RNA-stimulated hyper-phosphorylation of tristetraprolin/ZFP36 and AU-rich mRNA stabilization, Immunology, doi:10.1111/imm.13835.
14.
Yan et al., Super-resolution imaging reveals the mechanism of endosomal acidification inhibitors against SARS-CoV-2 infection, ChemBioChem, doi:10.1002/cbic.202400404.
15.
Mohd Abd Razak et al., In Vitro Anti-SARS-CoV-2 Activities of Curcumin and Selected Phenolic Compounds, Natural Product Communications, doi:10.1177/1934578X231188861.
16.
Alsmadi et al., The In Vitro, In Vivo, and PBPK Evaluation of a Novel Lung-Targeted Cardiac-Safe Hydroxychloroquine Inhalation Aerogel, AAPS PharmSciTech, doi:10.1208/s12249-023-02627-3.
17.
Wen et al., Cholinergic α7 nAChR signaling suppresses SARS-CoV-2 infection and inflammation in lung epithelial cells, Journal of Molecular Cell Biology, doi:10.1093/jmcb/mjad048.
18.
Kamga Kapchoup et al., In vitro effect of hydroxychloroquine on pluripotent stem cells and their cardiomyocytes derivatives, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1128382.
19.
Milan Bonotto et al., Cathepsin inhibitors nitroxoline and its derivatives inhibit SARS-CoV-2 infection, Antiviral Research, doi:10.1016/j.antiviral.2023.105655.
20.
Miao et al., SIM imaging resolves endocytosis of SARS-CoV-2 spike RBD in living cells, Cell Chemical Biology, doi:10.1016/j.chembiol.2023.02.001.
21.
Yuan et al., Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture, Communications Biology, doi:10.1038/s42003-022-03841-8.
22.
Faísca et al., Enhanced In Vitro Antiviral Activity of Hydroxychloroquine Ionic Liquids against SARS-CoV-2, Pharmaceutics, doi:10.3390/pharmaceutics14040877.
23.
Delandre et al., Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants, Pharmaceuticals, doi:10.3390/ph15040445.
24.
Purwati et al., An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia, PLOS One, doi:10.1371/journal.pone.0252302.
25.
Zhang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death & Differentiation, doi:10.1038/s41418-021-00782-3.
26.
Dang et al., Structural basis of anti-SARS-CoV-2 activity of hydroxychloroquine: specific binding to NTD/CTD and disruption of LLPS of N protein, bioRxiv, doi:10.1101/2021.03.16.435741.
27.
Shang (B) et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virology Journal, doi:10.1186/s12985-021-01515-1.
28.
Wang et al., Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus, Phytomedicine, doi:10.1016/j.phymed.2020.153333.
29.
Sheaff, R., A New Model of SARS-CoV-2 Infection Based on (Hydroxy)Chloroquine Activity, bioRxiv, doi:10.1101/2020.08.02.232892.
30.
Ou et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLOS Pathogens, doi:10.1371/journal.ppat.1009212.
31.
Andreani et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microbial Pathogenesis, doi:10.1016/j.micpath.2020.104228.
32.
Clementi et al., Combined Prophylactic and Therapeutic Use Maximizes Hydroxychloroquine Anti-SARS-CoV-2 Effects in vitro, Front. Microbiol., 10 July 2020, doi:10.3389/fmicb.2020.01704.
33.
Liu et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discovery 6, 16 (2020), doi:10.1038/s41421-020-0156-0.
34.
Yao et al., In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Clin. Infect. Dis., 2020 Mar 9, doi:10.1093/cid/ciaa237.
Bobrowski et al., 28 Feb 2021, USA, peer-reviewed, 13 authors.
Contact: murik@email.unc.edu, alexey.zakharov@nih.gov.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Synergistic and Antagonistic Drug Combinations against SARS-CoV-2
Molecular Therapy, doi:10.1016/j.ymthe.2020.12.016
Antiviral drug development for coronavirus disease 2019 (COVID-19) is occurring at an unprecedented pace, yet there are still limited therapeutic options for treating this disease. We hypothesized that combining drugs with independent mechanisms of action could result in synergy against SARS-CoV-2, thus generating better antiviral efficacy. Using in silico approaches, we prioritized 73 combinations of 32 drugs with potential activity against SARS-CoV-2 and then tested them in vitro. Sixteen synergistic and eight antagonistic combinations were identified; among 16 synergistic cases, combinations of the US Food and Drug Administration (FDA)-approved drug nitazoxanide with remdesivir, amodiaquine, or umifenovir were most notable, all exhibiting significant synergy against SARS-CoV-2 in a cell model. However, the combination of remdesivir and lysosomotropic drugs, such as hydroxychloroquine, demonstrated strong antagonism. Overall, these results highlight the utility of drug repurposing and preclinical testing of drug combinations for discovering potential therapies to treat COVID-19.
SUPPLEMENTAL INFORMATION Supplemental Information can be found online at https://doi.org/10. 1016/j.ymthe.2020.12.016 .
ACKNOWLEDGMENTS Data-mining tools used in this study were developed under the Biomedical Data Translator Initiative of the National Center for Advancing Translational Sciences, National Institutes of Health (NIH) (grants OT3TR002020 and OT2R002514) and under support of the NIH (grant 1U01CA207160). This research was also supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences (NCATS), NIH, United States.
DECLARATION OF INTERESTS The authors declare no competing interests.
References
Alves, Bobrowski, Melo-Filho, Korn, Auerbach et al., QSAR modeling of SARS-CoV Mpro inhibitors identifies Sufugolix, Cenicriviroc, Proglumetacin and other drugs as candidates for repurposing against SARS-CoV-2, Mol. Inform, doi:10.1002/minf.202000113
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the Treatment of Covid-19-Preliminary Report, N. Engl. J. Med
Bizon, Cox, Balhoff, Kebede, Wang et al., ROBOKOP KG and KGB: Integrated Knowledge Graphs from Federated Sources, J. Chem. Inf. Model
Brimacombe, Zhao, Eastman, Hu, Wang et al., An OpenData portal to share COVID-19 drug repurposing data in real time, bioRxiv, doi:10.1101/2020.06.04.135046
Bulusu, Guha, Mason, Lewis, Muratov et al., Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives, Drug Discov. Today
Cao, Wang, Wen, Liu, Wang et al., A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19, N. Engl. J. Med
Capuzzi, Thornton, Liu, Baker, Lam et al., Chemotext: A Publicly Available Web Server for Mining Drug-Target-Disease Relationships in PubMed, J. Chem. Inf. Model
Cherkasov, Muratov, Fourches, Varnek, Baskin et al., QSAR modeling: where have you been? Where are you going to?, J. Med. Chem
Chou, Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies, Pharmacol. Rev
Choudhary, Silakari, Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19, Virus Res
Davidson, Williamson, Lewis, Shoemark, Carroll et al., Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein, Genome Med
Deng, Zhong, Yu, Zhang, Wang et al., Pharmacokinetics, metabolism, and excretion of the antiviral drug arbidol in humans, Antimicrob. Agents Chemother
Eastman, Roth, Brimacombe, Simeonov, Shen et al., Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19, ACS Cent. Sci
Einav, Sobol, Gehrig, Glenn, The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors, J. Infect. Dis
Ferner, Aronson, Chloroquine and hydroxychloroquine in covid-19, BMJ
Foucquier, Guedj, Analysis of drug combinations: current methodological landscape, Pharmacol. Res. Perspect
Fourches, Muratov, Tropsha, Curation of chemogenomics data, Nat. Chem. Biol
Fourches, Muratov, Tropsha, Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation, J. Chem. Inf. Model
Fourches, Muratov, Tropsha, Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research, J. Chem. Inf. Model
Glaumann, Motakefi, Jansson, Intracellular distribution and effect of the antimalarial drug mefloquine on lysosomes of rat liver, Liver
Golbraikh, Tropsha, Beware of q2!, J. Mol. Graph. Model
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Hung, Lung, Tso, Liu, Chung et al., Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial, Lancet
Jasenosky, Cadena, Mire, Borisevich, Haridas et al., The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus, iScience
Johnson, Xie, Kalveram, Lokugamage, Muruato et al., Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis, bioRxiv, doi:10.1101/2020.08.26.268854
Jurgeit, Mcdowell, Moese, Meldrum, Schwendener et al., Niclosamide Is a Proton Carrier and Targets Acidic Endosomes with Broad Antiviral Effects, PLoS Pathog
Klimstra, Tilston-Lunel, Nambulli, Boslett, Mcmillen et al., SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients, J. Gen. Virol
Ko, Jeon, Ryu, Kim, Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells: Nafamostat is the most potent antiviral drug candidate, bioRxiv, doi:10.1101/2020.05.12.090035
Korn, Bobrowski, Li, Kebede, Wang et al., COVID-KOP: Integrating Emerging COVID-19 Data with the ROBOKOP Database, doi:10.26434/chemrxiv.12462623.V1
Menden, Wang, Mason, Szalai, Bulusu et al., Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen, Nat. Commun
Morton, Wang, Bizon, Cox, Balhoff et al., ROBOKOP: an abstraction layer and user interface for knowledge graphs to support question answering, Bioinformatics
Murakami, Wang, Babusis, Lepist, Sauer et al., Metabolism and pharmacokinetics of the anti-hepatitis C virus nucleotide prodrug GS-6620, Antimicrob. Agents Chemother
Muratov, Bajorath, Sheridan, Tetko, Filimonov et al., QSAR without borders, Chem. Soc. Rev
Muratov, Varlamova, Artemenko, Polishchuk, Kuz'min, Existing and Developing Approaches for QSAR Analysis of Mixtures, Mol. Inform
Muratov, Zakharov, Viribus Unitis: Drug Combinations as a Treatment Against COVID-19, chemRxiv, doi:10.26434/chemrxiv.12143355.v1
Rajoli, Pertinez, Arshad, Box, Tatham et al., Dose prediction for repurposing nitazoxanide in SARS-CoV-2 treatment or chemoprophylaxis, medRxiv, doi:10.1101/2020.05.01.20087130
Richards, Schwartz, Honeywell, Stewart, Cruz-Gordillo et al., Drug antagonism and single-agent dominance result from differences in death kinetics, Nat. Chem. Biol
Riva, Yuan, Yin, Martin-Sancho, Matsunaga et al., Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing, Nature
Rocco, Silva, Cruz, Junior, Tierno et al., Early use of nitazoxanide in mild Covid-19 disease: randomized, placebo-controlled trial, medRxiv, doi:10.1101/2020.10.21.20217208
Rossignol, Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus, J. Infect. Public Health
Stockis, Allemon, De Bruyn, Gengler, Nitazoxanide pharmacokinetics and tolerability in man using single ascending oral doses, Int. J. Clin. Pharmacol. Ther
Sun, He, Martínez-Romero, Kouznetsova, Tawa et al., Synergistic drug combination effectively blocks Ebola virus infection, Antiviral Res
Sun, Vilar, Tatonetti, High-Throughput Methods for Combinatorial Drug Discovery, Sci. Transl. Med
Tropsha, Best Practices for QSAR Model Development, Validation, and Exploitation, Mol. Inform
Vankadari, Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein, Int. J. Antimicrob. Agents
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res
Zakharov, Varlamova, Lagunin, Dmitriev, Muratov et al., QSAR Modeling and Prediction of Drug-Drug Interactions, Mol. Pharm
Zakharov, Zhao, Nguyen, Peryea, Sheils et al., Novel Consensus Architecture To Improve Performance of Large-Scale Multitask Deep Learning QSAR Models, J. Chem. Inf. Model
DOI record:
{
"DOI": "10.1016/j.ymthe.2020.12.016",
"ISSN": [
"1525-0016"
],
"URL": "http://dx.doi.org/10.1016/j.ymthe.2020.12.016",
"alternative-id": [
"S1525001620306730"
],
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Synergistic and Antagonistic Drug Combinations against SARS-CoV-2"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Molecular Therapy"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.ymthe.2020.12.016"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2020 The American Society of Gene and Cell Therapy."
}
],
"author": [
{
"affiliation": [],
"family": "Bobrowski",
"given": "Tesia",
"sequence": "first"
},
{
"affiliation": [],
"family": "Chen",
"given": "Lu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Eastman",
"given": "Richard T.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Itkin",
"given": "Zina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shinn",
"given": "Paul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Catherine Z.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Guo",
"given": "Hui",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zheng",
"given": "Wei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Michael",
"given": "Sam",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Simeonov",
"given": "Anton",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hall",
"given": "Matthew D.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zakharov",
"given": "Alexey V.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4616-7036",
"affiliation": [],
"authenticated-orcid": false,
"family": "Muratov",
"given": "Eugene N.",
"sequence": "additional"
}
],
"container-title": "Molecular Therapy",
"container-title-short": "Molecular Therapy",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"cell.com",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2020,
12,
16
]
],
"date-time": "2020-12-16T11:00:26Z",
"timestamp": 1608116426000
},
"deposited": {
"date-parts": [
[
2022,
2,
3
]
],
"date-time": "2022-02-03T20:47:56Z",
"timestamp": 1643921276000
},
"funder": [
{
"DOI": "10.13039/100000002",
"award": [
"1U01CA207160",
"OT2R002514",
"OT3TR002020"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000002",
"id-type": "DOI"
}
],
"name": "National Institutes of Health"
},
{
"DOI": "10.13039/100006108",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100006108",
"id-type": "DOI"
}
],
"name": "National Center for Advancing Translational Sciences"
}
],
"indexed": {
"date-parts": [
[
2025,
2,
21
]
],
"date-time": "2025-02-21T02:46:08Z",
"timestamp": 1740105968407,
"version": "3.37.3"
},
"is-referenced-by-count": 85,
"issue": "2",
"issued": {
"date-parts": [
[
2021,
2
]
]
},
"journal-issue": {
"issue": "2",
"published-print": {
"date-parts": [
[
2021,
2
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
2,
1
]
],
"date-time": "2021-02-01T00:00:00Z",
"timestamp": 1612137600000
}
},
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 367,
"start": {
"date-parts": [
[
2022,
2,
3
]
],
"date-time": "2022-02-03T00:00:00Z",
"timestamp": 1643846400000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S1525001620306730?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S1525001620306730?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "873-885",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2021,
2
]
]
},
"published-print": {
"date-parts": [
[
2021,
2
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1086/653080",
"article-title": "The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors",
"author": "Einav",
"doi-asserted-by": "crossref",
"first-page": "65",
"journal-title": "J. Infect. Dis.",
"key": "10.1016/j.ymthe.2020.12.016_bib1",
"volume": "202",
"year": "2010"
},
{
"DOI": "10.1016/j.antiviral.2016.11.017",
"article-title": "Synergistic drug combination effectively blocks Ebola virus infection",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "165",
"journal-title": "Antiviral Res.",
"key": "10.1016/j.ymthe.2020.12.016_bib2",
"volume": "137",
"year": "2017"
},
{
"DOI": "10.1124/pr.58.3.10",
"article-title": "Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies",
"author": "Chou",
"doi-asserted-by": "crossref",
"first-page": "621",
"journal-title": "Pharmacol. Rev.",
"key": "10.1016/j.ymthe.2020.12.016_bib3",
"volume": "58",
"year": "2006"
},
{
"DOI": "10.1126/scitranslmed.3006667",
"article-title": "High-Throughput Methods for Combinatorial Drug Discovery",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "205rv1",
"journal-title": "Sci. Transl. Med",
"key": "10.1016/j.ymthe.2020.12.016_bib4",
"volume": "5",
"year": "2013"
},
{
"DOI": "10.1016/j.virusres.2020.198146",
"article-title": "Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19",
"author": "Choudhary",
"doi-asserted-by": "crossref",
"first-page": "198146",
"journal-title": "Virus Res.",
"key": "10.1016/j.ymthe.2020.12.016_bib5",
"volume": "289",
"year": "2020"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105998",
"article-title": "Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein",
"author": "Vankadari",
"doi-asserted-by": "crossref",
"first-page": "105998",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "10.1016/j.ymthe.2020.12.016_bib6",
"volume": "56",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "crossref",
"first-page": "459",
"journal-title": "Nature",
"key": "10.1016/j.ymthe.2020.12.016_bib7",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1038/s41467-019-09799-2",
"article-title": "Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen",
"author": "Menden",
"doi-asserted-by": "crossref",
"first-page": "2674",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.ymthe.2020.12.016_bib8",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1016/S0140-6736(20)31042-4",
"article-title": "Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial",
"author": "Hung",
"doi-asserted-by": "crossref",
"first-page": "1695",
"journal-title": "Lancet",
"key": "10.1016/j.ymthe.2020.12.016_bib9",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2001282",
"article-title": "A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19",
"author": "Cao",
"doi-asserted-by": "crossref",
"first-page": "1787",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.ymthe.2020.12.016_bib10",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the Treatment of Covid-19—Preliminary Report",
"author": "Beigel",
"doi-asserted-by": "crossref",
"first-page": "1813",
"journal-title": "N. Engl. J. Med",
"key": "10.1016/j.ymthe.2020.12.016_bib11",
"volume": "383",
"year": "2020"
},
{
"article-title": "Viribus Unitis: Drug Combinations as a Treatment Against COVID-19",
"author": "Muratov",
"journal-title": "chemRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib12",
"year": "2020"
},
{
"DOI": "10.1038/s41589-020-0510-4",
"article-title": "Drug antagonism and single-agent dominance result from differences in death kinetics",
"author": "Richards",
"doi-asserted-by": "crossref",
"first-page": "791",
"journal-title": "Nat. Chem. Biol.",
"key": "10.1016/j.ymthe.2020.12.016_bib13",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1111/j.1600-0676.1992.tb01045.x",
"article-title": "Intracellular distribution and effect of the antimalarial drug mefloquine on lysosomes of rat liver",
"author": "Glaumann",
"doi-asserted-by": "crossref",
"first-page": "183",
"journal-title": "Liver",
"key": "10.1016/j.ymthe.2020.12.016_bib14",
"volume": "12",
"year": "1992"
},
{
"DOI": "10.1021/acscentsci.0c00489",
"article-title": "Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19",
"author": "Eastman",
"doi-asserted-by": "crossref",
"first-page": "672",
"journal-title": "ACS Cent. Sci.",
"key": "10.1016/j.ymthe.2020.12.016_bib15",
"volume": "6",
"year": "2020"
},
{
"DOI": "10.1128/AAC.02350-13",
"article-title": "Metabolism and pharmacokinetics of the anti-hepatitis C virus nucleotide prodrug GS-6620",
"author": "Murakami",
"doi-asserted-by": "crossref",
"first-page": "1943",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "10.1016/j.ymthe.2020.12.016_bib16",
"volume": "58",
"year": "2014"
},
{
"DOI": "10.1136/bmj.m1432",
"article-title": "Chloroquine and hydroxychloroquine in covid-19",
"author": "Ferner",
"doi-asserted-by": "crossref",
"first-page": "m1432",
"journal-title": "BMJ",
"key": "10.1016/j.ymthe.2020.12.016_bib17",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1016/j.jiph.2016.04.001",
"article-title": "Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus",
"author": "Rossignol",
"doi-asserted-by": "crossref",
"first-page": "227",
"journal-title": "J. Infect. Public Health",
"key": "10.1016/j.ymthe.2020.12.016_bib18",
"volume": "9",
"year": "2016"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "269",
"journal-title": "Cell Res.",
"key": "10.1016/j.ymthe.2020.12.016_bib19",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1016/j.isci.2019.07.003",
"article-title": "The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus",
"author": "Jasenosky",
"doi-asserted-by": "crossref",
"first-page": "1279",
"journal-title": "iScience",
"key": "10.1016/j.ymthe.2020.12.016_bib20",
"volume": "19",
"year": "2019"
},
{
"DOI": "10.1371/journal.ppat.1002976",
"article-title": "Niclosamide Is a Proton Carrier and Targets Acidic Endosomes with Broad Antiviral Effects",
"author": "Jurgeit",
"doi-asserted-by": "crossref",
"first-page": "e1002976",
"journal-title": "PLoS Pathog",
"key": "10.1016/j.ymthe.2020.12.016_bib21",
"volume": "8",
"year": "2012"
},
{
"article-title": "Dose prediction for repurposing nitazoxanide in SARS-CoV-2 treatment or chemoprophylaxis",
"author": "Rajoli",
"journal-title": "medRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib22",
"year": "2020"
},
{
"DOI": "10.5414/CPP40213",
"article-title": "Nitazoxanide pharmacokinetics and tolerability in man using single ascending oral doses",
"author": "Stockis",
"doi-asserted-by": "crossref",
"first-page": "213",
"journal-title": "Int. J. Clin. Pharmacol. Ther.",
"key": "10.1016/j.ymthe.2020.12.016_bib23",
"volume": "40",
"year": "2002"
},
{
"article-title": "Early use of nitazoxanide in mild Covid-19 disease: randomized, placebo-controlled trial",
"author": "Rocco",
"journal-title": "medRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib24",
"year": "2020"
},
{
"article-title": "Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells: Nafamostat is the most potent antiviral drug candidate",
"author": "Ko",
"journal-title": "bioRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib25",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2577-1",
"article-title": "Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing",
"author": "Riva",
"doi-asserted-by": "crossref",
"first-page": "113",
"journal-title": "Nature",
"key": "10.1016/j.ymthe.2020.12.016_bib26",
"volume": "586",
"year": "2020"
},
{
"DOI": "10.1186/s13073-020-00763-0",
"article-title": "Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein",
"author": "Davidson",
"doi-asserted-by": "crossref",
"first-page": "68",
"journal-title": "Genome Med.",
"key": "10.1016/j.ymthe.2020.12.016_bib27",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1099/jgv.0.001481",
"article-title": "SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients",
"author": "Klimstra",
"doi-asserted-by": "crossref",
"first-page": "1156",
"journal-title": "J. Gen. Virol.",
"key": "10.1016/j.ymthe.2020.12.016_bib28",
"volume": "101",
"year": "2020"
},
{
"article-title": "Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis",
"author": "Johnson",
"journal-title": "bioRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib29",
"year": "2020"
},
{
"DOI": "10.1128/AAC.02282-12",
"article-title": "Pharmacokinetics, metabolism, and excretion of the antiviral drug arbidol in humans",
"author": "Deng",
"doi-asserted-by": "crossref",
"first-page": "1743",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "10.1016/j.ymthe.2020.12.016_bib30",
"volume": "57",
"year": "2013"
},
{
"DOI": "10.1021/acs.jcim.7b00589",
"article-title": "Chemotext: A Publicly Available Web Server for Mining Drug-Target-Disease Relationships in PubMed",
"author": "Capuzzi",
"doi-asserted-by": "crossref",
"first-page": "212",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib31",
"volume": "58",
"year": "2018"
},
{
"DOI": "10.1021/acs.jcim.9b00683",
"article-title": "ROBOKOP KG and KGB: Integrated Knowledge Graphs from Federated Sources",
"author": "Bizon",
"doi-asserted-by": "crossref",
"first-page": "4968",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib32",
"volume": "59",
"year": "2019"
},
{
"article-title": "COVID-KOP: Integrating Emerging COVID-19 Data with the ROBOKOP Database",
"author": "Korn",
"journal-title": "chemRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib33",
"year": "2020"
},
{
"DOI": "10.1002/minf.201000061",
"article-title": "Best Practices for QSAR Model Development, Validation, and Exploitation",
"author": "Tropsha",
"doi-asserted-by": "crossref",
"first-page": "476",
"journal-title": "Mol. Inform.",
"key": "10.1016/j.ymthe.2020.12.016_bib34",
"volume": "29",
"year": "2010"
},
{
"DOI": "10.1016/j.drudis.2015.09.003",
"article-title": "Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives",
"author": "Bulusu",
"doi-asserted-by": "crossref",
"first-page": "225",
"journal-title": "Drug Discov. Today",
"key": "10.1016/j.ymthe.2020.12.016_bib35",
"volume": "21",
"year": "2016"
},
{
"DOI": "10.1021/acs.molpharmaceut.5b00762",
"article-title": "QSAR Modeling and Prediction of Drug-Drug Interactions",
"author": "Zakharov",
"doi-asserted-by": "crossref",
"first-page": "545",
"journal-title": "Mol. Pharm.",
"key": "10.1016/j.ymthe.2020.12.016_bib36",
"volume": "13",
"year": "2016"
},
{
"DOI": "10.1093/bioinformatics/btz604",
"article-title": "ROBOKOP: an abstraction layer and user interface for knowledge graphs to support question answering",
"author": "Morton",
"doi-asserted-by": "crossref",
"first-page": "5382",
"journal-title": "Bioinformatics",
"key": "10.1016/j.ymthe.2020.12.016_bib37",
"volume": "35",
"year": "2019"
},
{
"DOI": "10.1111/cts.12592",
"article-title": "The Biomedical Data Translator Program: Conception, Culture, and Community",
"doi-asserted-by": "crossref",
"first-page": "91",
"journal-title": "Clin. Transl. Sci.",
"key": "10.1016/j.ymthe.2020.12.016_bib38",
"volume": "12",
"year": "2019"
},
{
"key": "10.1016/j.ymthe.2020.12.016_bib39",
"series-title": "An AI challenge with AI2, CZI, MSR, Georgetown, NIH, and the White House. Kaggle",
"year": "2020"
},
{
"DOI": "10.1021/acs.jcim.9b00526",
"article-title": "Novel Consensus Architecture To Improve Performance of Large-Scale Multitask Deep Learning QSAR Models",
"author": "Zakharov",
"doi-asserted-by": "crossref",
"first-page": "4613",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib40",
"volume": "59",
"year": "2019"
},
{
"article-title": "QSAR modeling of SARS-CoV Mpro inhibitors identifies Sufugolix, Cenicriviroc, Proglumetacin and other drugs as candidates for repurposing against SARS-CoV-2",
"author": "Alves",
"journal-title": "Mol. Inform",
"key": "10.1016/j.ymthe.2020.12.016_bib41",
"year": "2020"
},
{
"DOI": "10.1039/D0CS00098A",
"article-title": "QSAR without borders",
"author": "Muratov",
"doi-asserted-by": "crossref",
"first-page": "3525",
"journal-title": "Chem. Soc. Rev.",
"key": "10.1016/j.ymthe.2020.12.016_bib42",
"volume": "49",
"year": "2020"
},
{
"DOI": "10.1021/jm4004285",
"article-title": "QSAR modeling: where have you been? Where are you going to?",
"author": "Cherkasov",
"doi-asserted-by": "crossref",
"first-page": "4977",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.ymthe.2020.12.016_bib43",
"volume": "57",
"year": "2014"
},
{
"DOI": "10.1021/ci100176x",
"article-title": "Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research",
"author": "Fourches",
"doi-asserted-by": "crossref",
"first-page": "1189",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib44",
"volume": "50",
"year": "2010"
},
{
"DOI": "10.1021/acs.jcim.6b00129",
"article-title": "Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation",
"author": "Fourches",
"doi-asserted-by": "crossref",
"first-page": "1243",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib45",
"volume": "56",
"year": "2016"
},
{
"DOI": "10.1038/nchembio.1881",
"article-title": "Curation of chemogenomics data",
"author": "Fourches",
"doi-asserted-by": "crossref",
"first-page": "535",
"journal-title": "Nat. Chem. Biol.",
"key": "10.1016/j.ymthe.2020.12.016_bib46",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1016/S1093-3263(01)00123-1",
"article-title": "Beware of q2!",
"author": "Golbraikh",
"doi-asserted-by": "crossref",
"first-page": "269",
"journal-title": "J. Mol. Graph. Model.",
"key": "10.1016/j.ymthe.2020.12.016_bib47",
"volume": "20",
"year": "2002"
},
{
"DOI": "10.1002/minf.201100129",
"article-title": "Existing and Developing Approaches for QSAR Analysis of Mixtures",
"author": "Muratov",
"doi-asserted-by": "crossref",
"first-page": "202",
"journal-title": "Mol. Inform.",
"key": "10.1016/j.ymthe.2020.12.016_bib48",
"volume": "31",
"year": "2012"
},
{
"article-title": "An OpenData portal to share COVID-19 drug repurposing data in real time",
"author": "Brimacombe",
"journal-title": "bioRxiv",
"key": "10.1016/j.ymthe.2020.12.016_bib49",
"year": "2020"
},
{
"DOI": "10.1002/prp2.149",
"article-title": "Analysis of drug combinations: current methodological landscape",
"author": "Foucquier",
"doi-asserted-by": "crossref",
"first-page": "e00149",
"journal-title": "Pharmacol. Res. Perspect.",
"key": "10.1016/j.ymthe.2020.12.016_bib50",
"volume": "3",
"year": "2015"
}
],
"reference-count": 50,
"references-count": 50,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2020.06.29.178889",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S1525001620306730"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Synergistic and Antagonistic Drug Combinations against SARS-CoV-2",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "29"
}
bobrowski