Identifying the Most Potent Dual-Targeting Compound(s) against 3CLprotease and NSP15exonuclease of SARS-CoV-2 from Nigella sativa: Virtual Screening via Physicochemical Properties, Docking and Dynamic Simulation Analysis
et al., Processes, doi:10.3390/pr9101814, Oct 2021
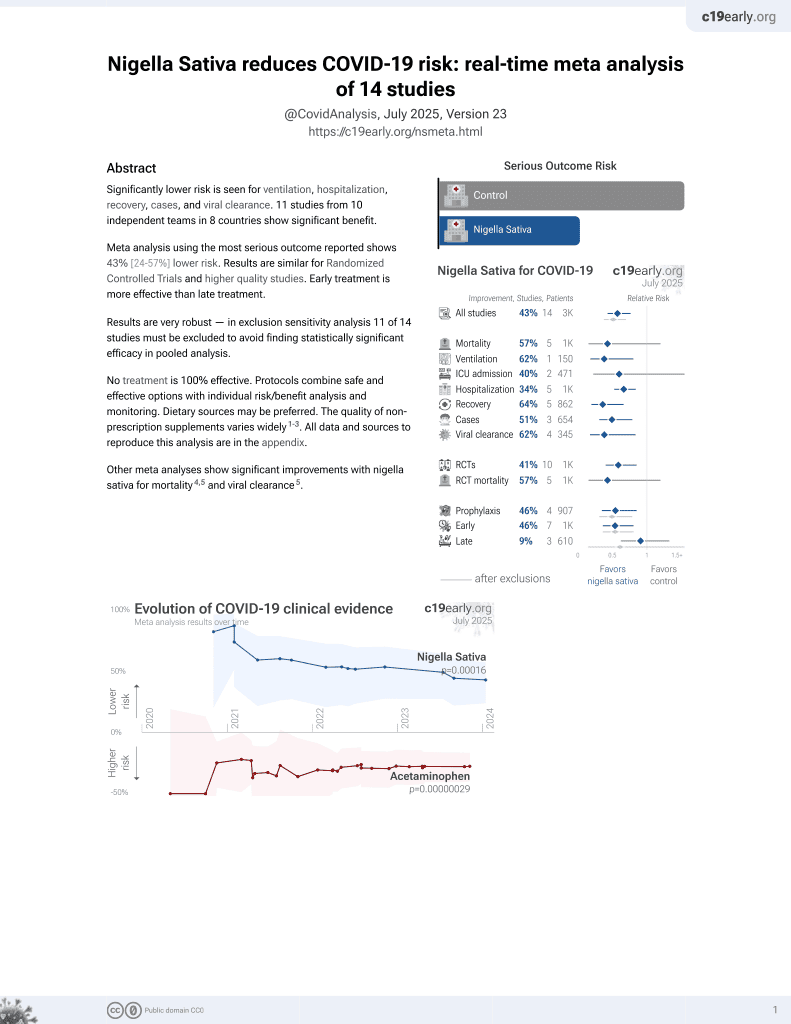
14th treatment shown to reduce risk in
January 2021, now with p = 0.00016 from 14 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
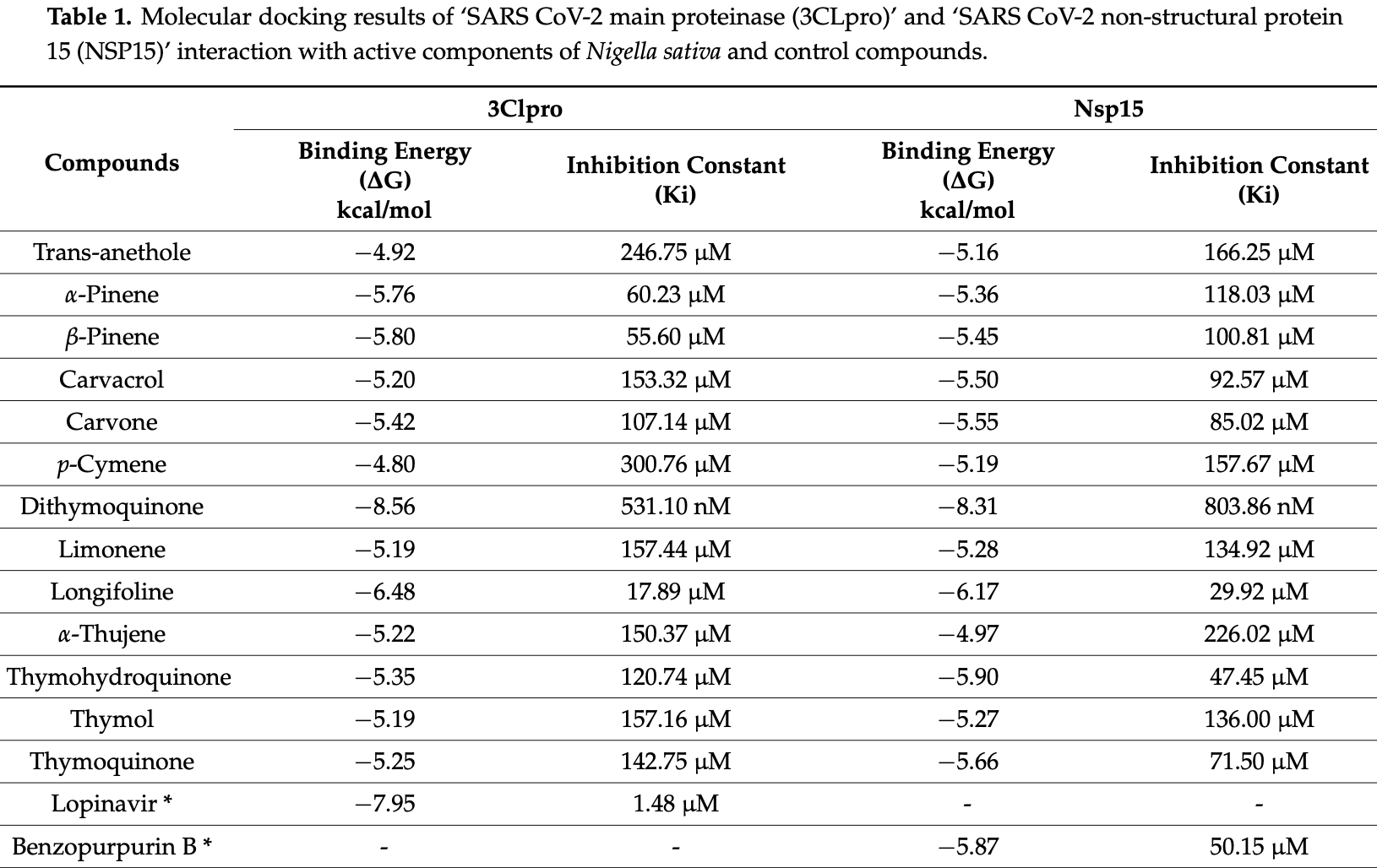
In silico study predicting that nigella sativa component dithymoquinone has good affinity and stability against SARS-CoV-2 3CLpro and NSP15, suggesting potential benefits for COVID-19 treatment.
22 preclinical studies support the efficacy of nigella sativa for COVID-19:
1.
Rahman et al., In Silico Screening of Potential Drug Candidate against Chain a of Coronavirus Binding Protein from Major Nigella Bioactive Compounds, Asian Journal of Advanced Research and Reports, doi:10.9734/ajarr/2024/v18i7697.
2.
Zafar Nayak Snehasis, S., Molecular Docking to Discover Potential Bio-Extract Substitutes for Hydroxychloroquine against COVID-19 and Malaria, International Journal of Science and Research (IJSR), doi:10.21275/SR24323192940.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Ali et al., Computational Prediction of Nigella sativa Compounds as Potential Drug Agents for Targeting Spike Protein of SARS-CoV-2, Pakistan BioMedical Journal, doi:10.54393/pbmj.v6i3.853.
5.
Miraz et al., Nigelladine A among Selected Compounds from Nigella sativa Exhibits Propitious Interaction with Omicron Variant of SARS-CoV-2: An In Silico Study, International Journal of Clinical Practice, doi:10.1155/2023/9917306.
6.
Sherwani et al., Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies, Processes, doi:10.3390/pr10071346.
7.
Khan et al., Inhibitory effect of thymoquinone from Nigella sativa against SARS-CoV-2 main protease. An in-silico study, Brazilian Journal of Biology, doi:10.1590/1519-6984.25066.
8.
Esharkawy et al., In vitro Potential Antiviral SARS-CoV-19- Activity of Natural Product Thymohydroquinone and Dithymoquinone from Nigela sativia, Bioorganic Chemistry, doi:10.1016/j.bioorg.2021.105587.
9.
Banerjee et al., Nigellidine (Nigella sativa, black-cumin seed) docking to SARS CoV-2 nsp3 and host inflammatory proteins may inhibit viral replication/transcription, Natural Product Research, doi:10.1080/14786419.2021.2018430.
10.
Rizvi et al., Identifying the Most Potent Dual-Targeting Compound(s) against 3CLprotease and NSP15exonuclease of SARS-CoV-2 from Nigella sativa: Virtual Screening via Physicochemical Properties, Docking and Dynamic Simulation Analysis, Processes, doi:10.3390/pr9101814.
11.
Mir et al., Identification of SARS-CoV-2 RNA-dependent RNA polymerase inhibitors from the major phytochemicals of Nigella sativa: An in silico approach, Saudi Journal of Biological Sciences, doi:10.1016/j.sjbs.2021.09.002.
12.
Hardianto et al., Exploring the Potency of Nigella sativa Seed in Inhibiting SARS-CoV-2 Main Protease Using Molecular Docking and Molecular Dynamics Simulations, Indonesian Journal of Chemistry, doi:10.22146/ijc.65951.
13.
Maiti et al., Active-site Molecular docking of Nigellidine to nucleocapsid/Nsp2/Nsp3/MPro of COVID-19 and to human IL1R and TNFR1/2 may stop viral-growth/cytokine-flood, and the drug source Nigella sativa (black cumin) seeds show potent antioxidant role in experimental rats, Research Square, doi:10.21203/rs.3.rs-26464/v1.
14.
Duru et al., In silico identification of compounds from Nigella sativa seed oil as potential inhibitors of SARS-CoV-2 targets, Bulletin of the National Research Centre, doi:10.1186/s42269-021-00517-x.
15.
Bouchentouf et al., Identification of Compounds from Nigella Sativa as New Potential Inhibitors of 2019 Novel Coronasvirus (Covid-19): Molecular Docking Study, ChemRxiv, doi:10.26434/chemrxiv.12055716.v1.
16.
Ali (B) et al., In vitro inhibitory effect of Nigella sativa L. extracts on SARS-COV-2 spike protein-ACE2 interaction, Current Therapeutic Research, doi:10.1016/j.curtheres.2024.100759.
17.
Bostancıklıoğlu et al., Nigella sativa, Anthemis hyaline and Citrus sinensis extracts reduce SARS-CoV-2 replication by fluctuating Rho GTPase, PI3K-AKT, and MAPK/ERK pathways in HeLa-CEACAM1a cells, Gene, doi:10.1016/j.gene.2024.148366.
Rizvi et al., 13 Oct 2021, peer-reviewed, 10 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Identifying the Most Potent Dual-Targeting Compound(s) against 3CLprotease and NSP15exonuclease of SARS-CoV-2 from Nigella sativa: Virtual Screening via Physicochemical Properties, Docking and Dynamic Simulation Analysis
Processes, doi:10.3390/pr9101814
Background: The outbreak of the coronavirus (SARS-CoV-2) has drastically affected the human population and caused enormous economic deprivation. It belongs to the β-coronavirus family and causes various problems such as acute respiratory distress syndrome and has resulted in a global pandemic. Though various medications have been under trial for combating COVID-19, specific medicine for treating COVID-19 is unavailable. Thus, the current situation urgently requires effective treatment modalities. Nigella sativa, a natural herb with reported antiviral activity and various pharmacological properties, has been selected in the present study to identify a therapeutic possibility for treating COVID-19. Methods: The present work aimed to virtually screen the bioactive compounds of N. sativa based on the physicochemical properties and docking approach against two SARS-CoV-2 enzymes responsible for crucial functions: 3CLpro (Main protease) and NSP15 (Nonstructural protein 15 or exonuclease). However, simulation trajectory analyses for 100 ns were accomplished by using the YASARA STRUCTURE tool based on the AMBER14 force field with 400 snapshots every 250 ps. RMSD and RMSF plots were successfully obtained for each target. Results: The results of molecular docking have shown higher binding energy of dithymoquinone (DTQ), a compound of N. sativa against 3CLpro and Nsp15, i.e., −8.56 kcal/mol and −8.31 kcal/mol, respectively. Further, the dynamic simulation has shown good stability of DTQ against both the targeted enzymes. In addition, physicochemical evaluation and toxicity assessment also revealed that DTQ obeyed the Lipinski rule and did not have any toxic side effects. Importantly, DTQ was much better in every aspect among the 13 N. sativa compounds and 2 control compounds tested. Conclusions: The results predicted that DTQ is a potent therapeutic molecule that could dual-target both 3CLpro and NSP15 for anti-COVID therapy.
References
Ahmad, Husain, Mujeeb, Khan, Najmi et al., A review on therapeutic potential of Nigella sativa: A miracle herb, Asian Pac. J. Trop. Biomed, doi:10.1016/S2221-1691(13)60075-1
Angeletti, Benvenuto, Bianchi, Giovanetti, Pascarella et al., COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis, J. Med. Virol, doi:10.1002/jmv.25719
Astani, Reichling, Schnitzler, Screening for antiviral activities of isolated compounds from essential oils, Evid.-Based Complementary Altern. Med, doi:10.1093/ecam/nep187
Badary, Hamza, Tikamdas, Thymoquinone: A Promising Natural Compound with Potential Benefits for COVID-19 Prevention and Cure, Drug Des. Dev. Ther, doi:10.2147/DDDT.S308863
Barakat, Shoman, Dina, Alfarouk, Antiviral activity and mode of action of Dianthus caryophyllus L. and Lupinus termes L. seed extracts against in vitro herpes simplex and hepatitis A viruses infection, J. Microbiol. Antimicrob
Berendsen, Postma, Van Gunsteren, Dinola, Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys, doi:10.1063/1.448118
Boopathi, Poma, Kolandaivel, Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1758788
Cascella, Rajnik, Cuomo, Dulebohn, Di Napoli et al., Evaluation, and Treatment of Coronavirus (COVID-19)
Chakravarty, Inhibition of histamine release from mast cells by nigellone, Ann Allergy Asthma Immunol
Chan, Kok, Zhu, Chu, To et al., Genomic characterization of the 2019 novel humanpathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan, Emerg. Microbes Infect, doi:10.1080/22221751.2020.1719902
Covid-, 19
Deng, Hackbart, Mettelman, O'brien, Mielech et al., Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1618310114
Dhama, Khan, Tiwari, Sircar, Bhat et al., Coronavirus Disease 2019-COVID-19, Clin. Microbiol. Rev, doi:10.1128/CMR.00028-20
Dorra, El-Barrawy, Sallam, Mahmoud, Evaluation of Antiviral and Antioxidant Activity of Selected Herbal Extracts, J. High Inst. Public Health, doi:10.21608/jhiph.2019.29464
El-Dakhakhny, Madi, Lembert, Ammon, Nigella sativa oil, nigellone and derived thymoquinone inhibit synthesis of 5-lipoxygenase products in polymorphonuclear leukocytes from rats, J. Ethnopharmacol, doi:10.1016/S0378-8741(02)00051-X
Faiza, Abdullah, Wang, Dithymoquinone as a novel inhibitor for 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) to prevent renal failure
Gavanji, Sayedipour, Larki, Bakhtari, Antiviral activity of some plant oils against herpes simplex virus type 1 in Vero cell culture, J. Acute Med, doi:10.1016/j.jacme.2015.07.001
Ghahramanloo, Kamalidehghan, Javar, Widodo, Majidzadeh et al., Comparative analysis of essential oil composition of Iranian and Indian Nigella sativa L. extracted using supercritical fluid extraction and solvent extraction, Drug Des. Dev. Ther, doi:10.2147/DDDT.S87251
Guo, Cao, Hong, Tan, Chen et al., The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-An update on the status, Mil. Med. Res, doi:10.1186/s40779-020-00240-0
Haddad, Picard, Bénard, Desvignes, Desprès et al., Ayapana triplinervis essential oil and its main component thymohydroquinone dimethyl ether inhibit Zika virus at doses devoid of toxicity in zebrafish, Molecules, doi:10.3390/molecules24193447
Haseena, Aithal, Das, Saheb, Phytochemical analysis of Nigella sativa and its effect on reproductive system, J. Pharm. Sci. Res
Hess, Bekker, Berendsen, Fraaije, LINCS: A linear constraint solver for molecular simulations, J. Comput. Chem, doi:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H
Islam, Parves, Paul, Uddin, Rahman et al., A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1761883
Jakalian, Jack, Bayly, Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation, J. Comput. Chem, doi:10.1002/jcc.10128
Jakhmola, Sehgal, Dogra, Saxena, Pande, Deciphering underlying mechanism of Sars-CoV-2 infection in humans and revealing the therapeutic potential of bioactive constituents from Nigella sativa to combat COVID19: In-Silico study, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1839560
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Keretsu, Bhujbal, Cho, Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation, Sci. Rep, doi:10.1038/s41598-020-74468-0
Kim, Jedrzejczak, Maltseva, Wilamowski, Endres et al., Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2, Protein Sci, doi:10.1002/pro.3873
Koshak, Koshak, Nigella, sativa L. as a potential phytotherapy for coronavirus disease 2019: A mini review of in silico studies, Curr. Ther. Res. Clin. Exp, doi:10.1016/j.curtheres.2020.100602
Krieger, Dunbrack, Jr, Hooft, Krieger, Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization, Methods Mol. Biol
Krieger, Vriend, New ways to boost molecular dynamics simulations, J. Comput. Chem, doi:10.1002/jcc.23899
Krieger, Vriend, YASARA View-Molecular graphics for all devices-From smartphones to workstations, Bioinformatics, doi:10.1093/bioinformatics/btu426
Lai, Chuang, Lee, Wei, Lin et al., Inhibition of herpes simplex virus type 1 by thymol-related monoterpenoids, Planta Med, doi:10.1055/s-0032-1315208
Lasarte-Cia, Lozano, Pérez-González, Gorraiz, Iribarren et al., Immunomodulatory properties of carvone inhalation and its effects on contextual fear memory in mice, Front. Immunol, doi:10.3389/fimmu.2018.00068
Laskowski, Swindells, LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery, J. Chem. Inf. Model, doi:10.1021/ci200227u
Lin, Hsu, Lin, Antiviral natural products and herbal medicines, J. Tradit. Complementary Med, doi:10.4103/2225-4110.124335
Lipinski, Lombardo, Dominy, Feeney, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Deliv. Rev, doi:10.1016/S0169-409X(00)00129-0
Lobo-Galo, Terrazas-López, Martínez-Martínez, Díaz-Sánchez, FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication, J. Biomol. Struct. Dyn
Maier, Martinez, Kasavajhala, Wickstrom, Hauser et al., ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput, doi:10.1021/acs.jctc.5b00255
Mcgonagle, Sharif, O'regan, Bridgewood, The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease, Autoimmun. Rev, doi:10.1016/j.autrev.2020.102537
Mirza, Froeyen, Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase, J. Pharm. Anal, doi:10.1016/j.jpha.2020.04.008
Molla, Azad, Al Hasib, Hossain, Ahammed et al., A review on antiviral effects of Nigella sativa L, Pharmacol. Online
Oyero, Toyama, Mitsuhiro, Onifade, Hidaka et al., Selective inhibition of hepatitis c virus replication by alpha-zam, a Nigella sativa seed formulation, Afr. J. Tradit. Complementary Altern. Med, doi:10.21010/ajtcam.v13i6.20
Rizvi, Shakil, Haneef, A simple click by click protocol to perform docking: AutoDock 4.2 made easy for nonbioinformaticians, EXCLI J
Ryabchenko, Tulupova, Schmidt, Wlcek, Buchbauer et al., Investigation of anticancer and antiviral properties of selected aroma samples, Nat. Prod. Commun, doi:10.1177/1934578X0800300710
Sahak, Kabir, Abbas, Draman, Hashim et al., The role of Nigella sativa and its active constituents in learning and memory, Evid.-Based Complementary Altern. Med, doi:10.1155/2016/6075679
Sharifi-Rad, Salehi, Schnitzler, Ayatollahi, Kobarfard et al., Susceptibility of herpes simplex virus type 1 to monoterpenes thymol, carvacrol, p-cymene and essential oils of Sinapis arvensis L., Lallemantia royleana Benth. and Pulicaria vulgaris Gaertn, Cell. Mol. Biol, doi:10.14715/cmb/2017.63.8.10
Sinha, Shakya, Prasad, Singh, Gurav et al., An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1762741
Song, Xu, Bao, Zhang, Yu et al., From SARS to MERS, Thrusting Coronaviruses into the Spotlight, Viruses, doi:10.3390/v11010059
Toma, Simu, Hanganu, Olah, Vata et al., Chemical composition of the Tunisian Nigella sativa. Note I. Profile on essential oil, Farmacia
Wang, Wang, Wu, Xu, Li, Antiviral mechanism of carvacrol on HSV-2 infectivity through inhibition of RIP3-mediated programmed cell necrosis pathway and ubiquitin-proteasome system in BSC-1 cells, BMC Infect. Dis
Wang, Wolf, Caldwell, Kollman, Case, Development and testing of a general amber force field, J. Comput. Chem, doi:10.1002/jcc.20035
Wu, Chen, Cai, Xia, Zhou et al., Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China, JAMA Intern. Med, doi:10.1001/jamainternmed.2020.0994
Yang, Wu, Zu, Fu, Comparative anti-infectious bronchitis virus (IBV) activity of (−)-pinene: Effect on nucleocapsid (N) protein, Molecules, doi:10.3390/molecules16021044
DOI record:
{
"DOI": "10.3390/pr9101814",
"ISSN": [
"2227-9717"
],
"URL": "http://dx.doi.org/10.3390/pr9101814",
"abstract": "<jats:p>Background: The outbreak of the coronavirus (SARS-CoV-2) has drastically affected the human population and caused enormous economic deprivation. It belongs to the β-coronavirus family and causes various problems such as acute respiratory distress syndrome and has resulted in a global pandemic. Though various medications have been under trial for combating COVID-19, specific medicine for treating COVID-19 is unavailable. Thus, the current situation urgently requires effective treatment modalities. Nigella sativa, a natural herb with reported antiviral activity and various pharmacological properties, has been selected in the present study to identify a therapeutic possibility for treating COVID-19. Methods: The present work aimed to virtually screen the bioactive compounds of N. sativa based on the physicochemical properties and docking approach against two SARS-CoV-2 enzymes responsible for crucial functions: 3CLpro (Main protease) and NSP15 (Nonstructural protein 15 or exonuclease). However, simulation trajectory analyses for 100 ns were accomplished by using the YASARA STRUCTURE tool based on the AMBER14 force field with 400 snapshots every 250 ps. RMSD and RMSF plots were successfully obtained for each target. Results: The results of molecular docking have shown higher binding energy of dithymoquinone (DTQ), a compound of N. sativa against 3CLpro and Nsp15, i.e., −8.56 kcal/mol and −8.31 kcal/mol, respectively. Further, the dynamic simulation has shown good stability of DTQ against both the targeted enzymes. In addition, physicochemical evaluation and toxicity assessment also revealed that DTQ obeyed the Lipinski rule and did not have any toxic side effects. Importantly, DTQ was much better in every aspect among the 13 N. sativa compounds and 2 control compounds tested. Conclusions: The results predicted that DTQ is a potent therapeutic molecule that could dual-target both 3CLpro and NSP15 for anti-COVID therapy.</jats:p>",
"alternative-id": [
"pr9101814"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-3106-4707",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rizvi",
"given": "Syed Mohd Danish",
"sequence": "first"
},
{
"affiliation": [],
"family": "Hussain",
"given": "Talib",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3902-1162",
"affiliation": [],
"authenticated-orcid": false,
"family": "Moin",
"given": "Afrasim",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3966-7278",
"affiliation": [],
"authenticated-orcid": false,
"family": "Dixit",
"given": "Sheshagiri R.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mandal",
"given": "Subhankar P.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7080-6822",
"affiliation": [],
"authenticated-orcid": false,
"family": "Adnan",
"given": "Mohd",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5525-708X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Jamal",
"given": "Qazi Mohammad Sajid",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4003-565X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sharma",
"given": "Dinesh C.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Alanazi",
"given": "Abulrahman Sattam",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Unissa",
"given": "Rahamat",
"sequence": "additional"
}
],
"container-title": [
"Processes"
],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
10,
14
]
],
"date-time": "2021-10-14T01:48:39Z",
"timestamp": 1634176119000
},
"deposited": {
"date-parts": [
[
2021,
10,
21
]
],
"date-time": "2021-10-21T02:06:17Z",
"timestamp": 1634781977000
},
"funder": [
{
"DOI": "10.13039/501100008809",
"award": [
"RG-20 144"
],
"doi-asserted-by": "publisher",
"name": "University of Hail"
}
],
"indexed": {
"date-parts": [
[
2021,
10,
22
]
],
"date-time": "2021-10-22T09:50:38Z",
"timestamp": 1634896238898
},
"is-referenced-by-count": 0,
"issn-type": [
{
"type": "electronic",
"value": "2227-9717"
}
],
"issue": "10",
"issued": {
"date-parts": [
[
2021,
10,
13
]
]
},
"journal-issue": {
"issue": "10",
"published-online": {
"date-parts": [
[
2021,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
10,
13
]
],
"date-time": "2021-10-13T00:00:00Z",
"timestamp": 1634083200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2227-9717/9/10/1814/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1814",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2021,
10,
13
]
]
},
"published-online": {
"date-parts": [
[
2021,
10,
13
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1080/07391102.2020.1839560",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"key": "ref2",
"unstructured": "WHO COVID-19, WHO 2020https://www.who.int/health-topics/coronavirus#tab=tab_1"
},
{
"DOI": "10.1016/j.autrev.2020.102537",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"DOI": "10.1001/jamainternmed.2020.0994",
"doi-asserted-by": "publisher",
"key": "ref4"
},
{
"article-title": "Features, Evaluation, and Treatment of Coronavirus (COVID-19)",
"author": "Cascella",
"key": "ref5",
"series-title": "StatPearls [Internet]",
"year": "2021"
},
{
"DOI": "10.3390/v11010059",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1128/CMR.00028-20",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1186/s40779-020-00240-0",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1080/07391102.2020.1758788",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1016/j.jpha.2020.04.008",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1002/jmv.25719",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1080/22221751.2020.1719902",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.4103/2225-4110.124335",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1016/j.curtheres.2020.100602",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.1016/S2221-1691(13)60075-1",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"article-title": "A review on antiviral effects of Nigella sativa L.",
"author": "Molla",
"first-page": "47",
"journal-title": "Pharmacol. Online",
"key": "ref16",
"volume": "2",
"year": "2019"
},
{
"article-title": "Antiviral activity and mode of action of Dianthus caryophyllus L. and Lupinus termes L. seed extracts against in vitro herpes simplex and hepatitis A viruses infection",
"author": "Barakat",
"first-page": "23",
"journal-title": "J. Microbiol. Antimicrob.",
"key": "ref17",
"volume": "2",
"year": "2010"
},
{
"DOI": "10.21010/ajtcam.v13i6.20",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.21608/jhiph.2019.29464",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"article-title": "Chemical composition of the Tunisian Nigella sativa. Note I. Profile on essential oil",
"author": "Toma",
"first-page": "458",
"journal-title": "Farmacia",
"key": "ref20",
"volume": "58",
"year": "2010"
},
{
"DOI": "10.1155/2016/6075679",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.2147/DDDT.S87251",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"article-title": "Phytochemical analysis of Nigella sativa and its effect on reproductive system",
"author": "Haseena",
"first-page": "514",
"journal-title": "J. Pharm. Sci. Res.",
"key": "ref23",
"volume": "7",
"year": "2015"
},
{
"DOI": "10.1093/ecam/nep187",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"DOI": "10.3390/molecules16021044",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1186/s12879-020-05556-9",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.3389/fimmu.2018.00068",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.14715/cmb/2017.63.8.10",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1177/1934578X0800300710",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"DOI": "10.1016/j.jacme.2015.07.001",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.3390/molecules24193447",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.1055/s-0032-1315208",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.2147/DDDT.S308863",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"article-title": "Inhibition of histamine release from mast cells by nigellone",
"author": "Chakravarty",
"first-page": "237",
"journal-title": "Ann Allergy Asthma Immunol.",
"key": "ref34",
"volume": "70",
"year": "1993"
},
{
"DOI": "10.1016/S0378-8741(02)00051-X",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"article-title": "Dithymoquinone as a novel inhibitor for 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) to prevent renal failure",
"author": "Faiza",
"journal-title": "arXiv",
"key": "ref36",
"year": "2017"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1002/pro.3873",
"doi-asserted-by": "publisher",
"key": "ref38"
},
{
"key": "ref39",
"unstructured": "www.openmolecules.orghttp://www.openmolecules.org/datawarrior/download.html"
},
{
"DOI": "10.1016/S0169-409X(00)00129-0",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"article-title": "A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians",
"author": "Rizvi",
"first-page": "831",
"journal-title": "EXCLI J.",
"key": "ref41",
"volume": "12",
"year": "2013"
},
{
"DOI": "10.1021/ci200227u",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.1093/bioinformatics/btu426",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"DOI": "10.1007/978-1-61779-465-0_25",
"article-title": "Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization",
"author": "Krieger",
"doi-asserted-by": "crossref",
"first-page": "405",
"journal-title": "Methods Mol. Biol.",
"key": "ref44",
"volume": "819",
"year": "2012"
},
{
"DOI": "10.1021/acs.jctc.5b00255",
"doi-asserted-by": "publisher",
"key": "ref45"
},
{
"DOI": "10.1002/jcc.10128",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1002/jcc.20035",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.1002/jcc.23899",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1063/1.448118",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.1038/s41598-020-74468-0",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"DOI": "10.1080/07391102.2020.1761883",
"doi-asserted-by": "publisher",
"key": "ref52"
},
{
"DOI": "10.1080/07391102.2020.1764393",
"doi-asserted-by": "publisher",
"key": "ref53"
},
{
"DOI": "10.1073/pnas.1618310114",
"doi-asserted-by": "publisher",
"key": "ref54"
},
{
"DOI": "10.1080/07391102.2020.1762741",
"doi-asserted-by": "publisher",
"key": "ref55"
}
],
"reference-count": 55,
"references-count": 55,
"relation": {},
"score": 1,
"short-container-title": [
"Processes"
],
"short-title": [],
"source": "Crossref",
"subject": [
"Process Chemistry and Technology",
"Chemical Engineering (miscellaneous)",
"Bioengineering"
],
"subtitle": [],
"title": [
"Identifying the Most Potent Dual-Targeting Compound(s) against 3CLprotease and NSP15exonuclease of SARS-CoV-2 from Nigella sativa: Virtual Screening via Physicochemical Properties, Docking and Dynamic Simulation Analysis"
],
"type": "journal-article",
"volume": "9"
}