Exploring the Potency of Nigella sativa Seed in Inhibiting SARS-CoV-2 Main Protease Using Molecular Docking and Molecular Dynamics Simulations
et al., Indonesian Journal of Chemistry, doi:10.22146/ijc.65951, Jul 2021
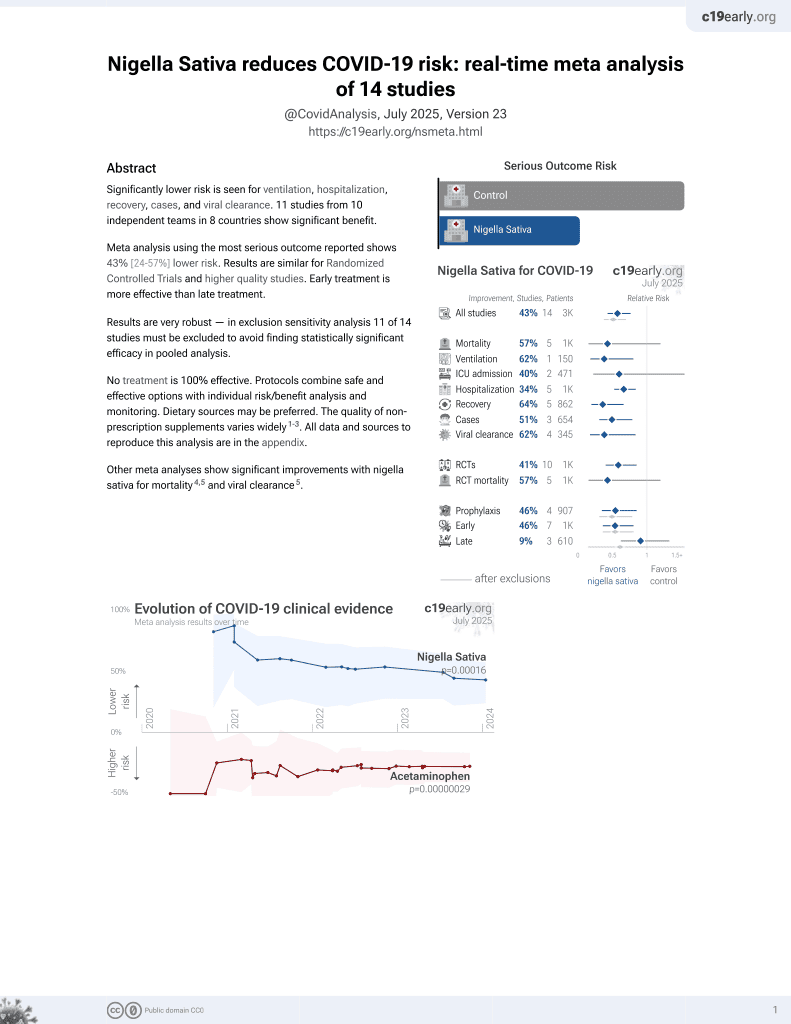
14th treatment shown to reduce risk in
January 2021, now with p = 0.00016 from 14 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
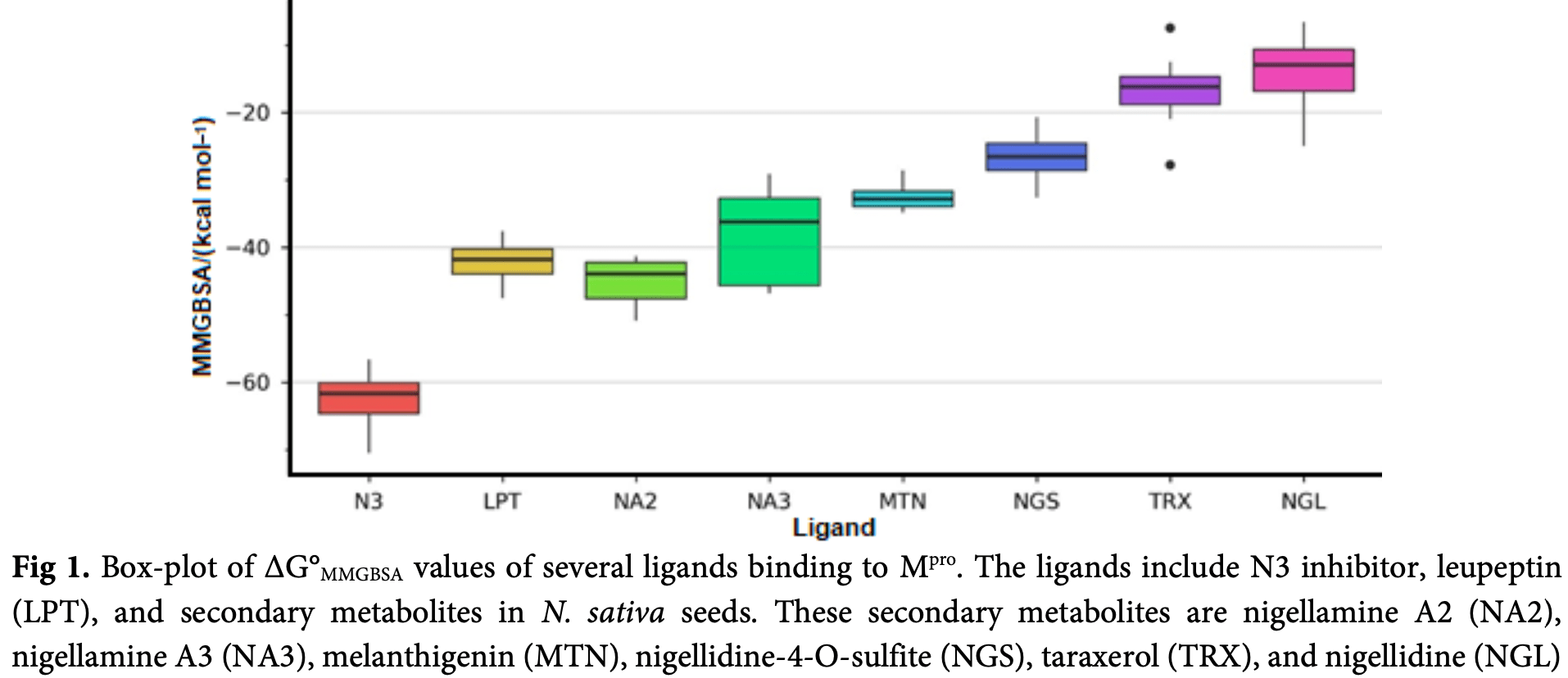
In silico study showing nigella sativa metabolites binding to SARS-CoV-2 Mpro.
22 preclinical studies support the efficacy of nigella sativa for COVID-19:
1.
Rahman et al., In Silico Screening of Potential Drug Candidate against Chain a of Coronavirus Binding Protein from Major Nigella Bioactive Compounds, Asian Journal of Advanced Research and Reports, doi:10.9734/ajarr/2024/v18i7697.
2.
Zafar Nayak Snehasis, S., Molecular Docking to Discover Potential Bio-Extract Substitutes for Hydroxychloroquine against COVID-19 and Malaria, International Journal of Science and Research (IJSR), doi:10.21275/SR24323192940.
3.
Alkafaas et al., A study on the effect of natural products against the transmission of B.1.1.529 Omicron, Virology Journal, doi:10.1186/s12985-023-02160-6.
4.
Ali et al., Computational Prediction of Nigella sativa Compounds as Potential Drug Agents for Targeting Spike Protein of SARS-CoV-2, Pakistan BioMedical Journal, doi:10.54393/pbmj.v6i3.853.
5.
Miraz et al., Nigelladine A among Selected Compounds from Nigella sativa Exhibits Propitious Interaction with Omicron Variant of SARS-CoV-2: An In Silico Study, International Journal of Clinical Practice, doi:10.1155/2023/9917306.
6.
Sherwani et al., Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies, Processes, doi:10.3390/pr10071346.
7.
Khan et al., Inhibitory effect of thymoquinone from Nigella sativa against SARS-CoV-2 main protease. An in-silico study, Brazilian Journal of Biology, doi:10.1590/1519-6984.25066.
8.
Esharkawy et al., In vitro Potential Antiviral SARS-CoV-19- Activity of Natural Product Thymohydroquinone and Dithymoquinone from Nigela sativia, Bioorganic Chemistry, doi:10.1016/j.bioorg.2021.105587.
9.
Banerjee et al., Nigellidine (Nigella sativa, black-cumin seed) docking to SARS CoV-2 nsp3 and host inflammatory proteins may inhibit viral replication/transcription, Natural Product Research, doi:10.1080/14786419.2021.2018430.
10.
Rizvi et al., Identifying the Most Potent Dual-Targeting Compound(s) against 3CLprotease and NSP15exonuclease of SARS-CoV-2 from Nigella sativa: Virtual Screening via Physicochemical Properties, Docking and Dynamic Simulation Analysis, Processes, doi:10.3390/pr9101814.
11.
Mir et al., Identification of SARS-CoV-2 RNA-dependent RNA polymerase inhibitors from the major phytochemicals of Nigella sativa: An in silico approach, Saudi Journal of Biological Sciences, doi:10.1016/j.sjbs.2021.09.002.
12.
Hardianto et al., Exploring the Potency of Nigella sativa Seed in Inhibiting SARS-CoV-2 Main Protease Using Molecular Docking and Molecular Dynamics Simulations, Indonesian Journal of Chemistry, doi:10.22146/ijc.65951.
13.
Maiti et al., Active-site Molecular docking of Nigellidine to nucleocapsid/Nsp2/Nsp3/MPro of COVID-19 and to human IL1R and TNFR1/2 may stop viral-growth/cytokine-flood, and the drug source Nigella sativa (black cumin) seeds show potent antioxidant role in experimental rats, Research Square, doi:10.21203/rs.3.rs-26464/v1.
14.
Duru et al., In silico identification of compounds from Nigella sativa seed oil as potential inhibitors of SARS-CoV-2 targets, Bulletin of the National Research Centre, doi:10.1186/s42269-021-00517-x.
15.
Bouchentouf et al., Identification of Compounds from Nigella Sativa as New Potential Inhibitors of 2019 Novel Coronasvirus (Covid-19): Molecular Docking Study, ChemRxiv, doi:10.26434/chemrxiv.12055716.v1.
16.
Ali (B) et al., In vitro inhibitory effect of Nigella sativa L. extracts on SARS-COV-2 spike protein-ACE2 interaction, Current Therapeutic Research, doi:10.1016/j.curtheres.2024.100759.
17.
Bostancıklıoğlu et al., Nigella sativa, Anthemis hyaline and Citrus sinensis extracts reduce SARS-CoV-2 replication by fluctuating Rho GTPase, PI3K-AKT, and MAPK/ERK pathways in HeLa-CEACAM1a cells, Gene, doi:10.1016/j.gene.2024.148366.
Hardianto et al., 12 Jul 2021, peer-reviewed, 5 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Exploring the Potency of Nigella sativa Seed in Inhibiting SARS-CoV-2 Main Protease Using Molecular Docking and Molecular Dynamics Simulations
Indonesian Journal of Chemistry, doi:10.22146/ijc.65951
Coronavirus disease (COVID-19) is a pandemic burdening the global economy. It is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Black cumin (Nigella sativa) seed may contain antivirals for the disease since it was reported to inhibit the human immunodeficiency virus (HIV) and hepatitis C virus (HCV). Main protease (M pro ) is a vital protein for viral replication and a promising target for COVID-19 drug development. Hence, in this study, we intended to uncover the potency of N. sativa seed as the natural source of inhibitors for SARS-CoV-2 M pro . We collected secondary metabolites in N. sativa seed through a literature search and employed Lipinski's rule of five as the initial filter. Subsequently, virtual screening campaigns using a molecular docking method were performed, with N3 inhibitor and leupeptin as reference ligands. The top hits were analyzed further using a molecular dynamics simulation approach. Molecular dynamics simulations showed that binding affinities of nigellamine A2 and A3 to M pro are comparable to that of leupeptin, with median values of -43.9 and -36.2 kcal mol -1 , respectively. Ultimately, this study provides scientific information regarding N. sativa seeds' potency against COVID-19 and helps direct further wet experiments.
References
Ahmad, Husain, Mujeeb, Khan, Najmi et al., A review on therapeutic potential of Nigella sativa: A miracle herb, Asian Pac. J. Trop. Biomed
Ashraf, Ashraf, Ashraf, Imran, Kalsoom et al., Honey and Nigella sativa against COVID-19 in Pakistan (HNS-COVID-PK): A multi-center placebo-controlled randomized clinical trial, medRxiv
Bouchentouf, Missoum, Identification of compounds from Nigella Sativa as new potential inhibitors of 2019 novel Coronasvirus (Covid-19), Molecular docking study
Case, Aktulga, Belfon, Ben-Shalom, Brozell et al., Amber 2020
Egbert, Whitty, Keserű, Vajda, Why some targets benefit from beyond rule of five drugs, J. Med. Chem
Ferdian, Elfirta, Emilia, Ikhwani, Inhibitory potential of black seed (Nigella sativa L.) bioactive compounds towards main protease of SARS-CoV-2: In silico study, Ann. Bogor
Forli, Huey, Pique, Sanner, Goodsell et al., Computational protein-ligand docking and virtual drug screening with the AutoDock suite, Nat. Protoc
Frisch, Trucks, Schlegel, Scuseria, Robb et al., Gaussian 09 Revision A.02
Gorbalenya, Baker, Baric, De Groot, Drosten et al., The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol
Hardianto, Khanna, Liu, Ranganathan, Diverse dynamics features of novel protein kinase C (PKC) isozymes determine the selectivity of a fluorinated balanol analogue for PKCϵ, BMC Bioinf
Hardianto, Liu, Ranganathan, Molecular dynamics pinpoint the global fluorine effect in balanoids binding to PKCε and PKA, J. Chem. Inf. Model
Hardianto, Yusuf, Liu, Ranganathan, Exploration of charge states of balanol analogues acting as ATP-competitive inhibitors in kinases, BMC Bioinf
Jin, Du, Xu, Deng, Liu et al., Structure of M pro from SARS-CoV-2 and discovery of its inhibitors, Nature
Khan, Afzal, Chemical composition of Nigella sativa Linn: Part 2 Recent advances, Inflammopharmacology
Khan, Siddiqui, Jain, Sonwane, Discovery of potential inhibitors of SARS-CoV-2 (COVID-19) Main Protease (Mpro) from Nigella Sativa (black seed) by molecular docking study, Coronaviruses
Kneller, Galanie, Phillips, O'neill, Coates et al., Malleability of the SARS-CoV-2 3CL M pro activesite cavity facilitates binding of clinical antivirals, Structure
Lipinski, Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions, Adv. Drug Delivery Rev
Maier, Martinez, Kasavajhala, Wickstrom, Hauser et al., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB, J. Chem. Theory Comput
Maiti, Banerjee, Nazmeen, Kanwar, Das, Active-site molecular docking of Nigellidine with nucleocapsid-NSP2-M Pro of COVID-19 and to human IL1R-IL6R and strong antioxidant role of Nigella-sativa in experimental rats, J. Drug Targeting
Mathieu, Ritchie, Ortiz-Ospina, Roser, Hasell et al., A global database of COVID-19 vaccinations, Nat. Hum. Behav
Mehta, Pandit, Gupta, New principles from seeds of Nigella sativa, Nat. Prod. Res
Merfort, Wray, Barakat, Hussein, Nawwar et al., Flavonol triglycosides from seeds of Nigella sativa, Phytochemistry
Morikawa, Xu, Ninomiya, Matsuda, Yoshikawa, Nigellamines A3, A4, A5, and C, new dolabellane-type diterpene alkaloids, with lipid metabolism-promoting activities from the Egyptian medicinal food black cumin, Chem. Pharm. Bull
Nickavar, Mojab, Javidnia, Amoli, Chemical composition of the fixed and volatile oils of Nigella sativa L. from Iran, Z. Naturforsch
Simmons, Gosalia, Rennekamp, Reeves, Diamond et al., Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry, Proc. Natl. Acad. Sci. U.S.A
Singh, Villoutreix, Ecker, Rigorous sampling of docking poses unveils binding hypothesis for the halogenated ligands of L-type Amino acid Transporter 1 (LAT1), Sci. Rep
Stewart, Application of the PM6 method to modeling proteins, J. Mol. Model
Sumaryada, Pramudita, Molecular docking evaluation of some Indonesian's popular herbals for a possible COVID-19 treatment, Biointerface Res. Appl. Chem
Torres, Sodero, Jofily, Silva, Key topics in molecular docking for drug design, Int. J. Mol. Sci
Who, Pneumonia of unknown cause -China
Who, WHO announces COVID-19 outbreak a pandemic
Worldometers, COVID-19 coronavirus pandemic
Yimer, Tuem, Karim, Ur-Rehman, Anwar, Nigella sativa L. (black cumin): A promising natural remedy for wide range of illnesses, Evidence-Based Complementary, Altern. Med
Yuan, Nahar, Sharma, Liu, Slitt et al., Indazole-type alkaloids from Nigella sativa seeds exhibit antihyperglycemic effects via AMPK activation in vitro, J. Nat. Prod
Yusuf, Hardianto, Muchtaridi, Nuwarda, Subroto, Introduction of Docking-Based Virtual Screening Workflow Using Desktop Personal Computer
DOI record:
{
"DOI": "10.22146/ijc.65951",
"ISSN": [
"2460-1578",
"1411-9420"
],
"URL": "http://dx.doi.org/10.22146/ijc.65951",
"abstract": "<jats:p>Coronavirus disease (COVID-19) is a pandemic burdening the global economy. It is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Black cumin (Nigella sativa) seed may contain antivirals for the disease since it was reported to inhibit the human immunodeficiency virus (HIV) and hepatitis C virus (HCV). Main protease (Mpro) is a vital protein for viral replication and a promising target for COVID-19 drug development. Hence, in this study, we intended to uncover the potency of N. sativa seed as the natural source of inhibitors for SARS-CoV-2 Mpro. We collected secondary metabolites in N. sativa seed through a literature search and employed Lipinski’s rule of five as the initial filter. Subsequently, virtual screening campaigns using a molecular docking method were performed, with N3 inhibitor and leupeptin as reference ligands. The top hits were analyzed further using a molecular dynamics simulation approach. Molecular dynamics simulations showed that binding affinities of nigellamine A2 and A3 to Mpro are comparable to that of leupeptin, with median values of -43.9 and -36.2 kcal mol–1, respectively. Ultimately, this study provides scientific information regarding N. sativa seeds’ potency against COVID-19 and helps direct further wet experiments.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0001-6065-5437",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hardianto",
"given": "Ari",
"sequence": "first"
},
{
"affiliation": [],
"family": "Yusuf",
"given": "Muhammad",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hidayat",
"given": "Ika Wiani",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ishmayana",
"given": "Safri",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Soedjanaatmadja",
"given": "Ukun Mochammad Syukur",
"sequence": "additional"
}
],
"container-title": "Indonesian Journal of Chemistry",
"container-title-short": "Indones. J. Chem.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
10,
12
]
],
"date-time": "2021-10-12T06:50:24Z",
"timestamp": 1634021424000
},
"deposited": {
"date-parts": [
[
2021,
10,
12
]
],
"date-time": "2021-10-12T06:50:45Z",
"timestamp": 1634021445000
},
"indexed": {
"date-parts": [
[
2024,
5,
14
]
],
"date-time": "2024-05-14T06:31:30Z",
"timestamp": 1715668290613
},
"is-referenced-by-count": 2,
"issue": "5",
"issued": {
"date-parts": [
[
2021,
8,
31
]
]
},
"journal-issue": {
"issue": "5",
"published-online": {
"date-parts": [
[
2021,
8,
14
]
]
}
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
8,
31
]
],
"date-time": "2021-08-31T00:00:00Z",
"timestamp": 1630368000000
}
}
],
"link": [
{
"URL": "https://jurnal.ugm.ac.id/ijc/article/viewFile/65951/31947",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://jurnal.ugm.ac.id/ijc/article/viewFile/65951/31948",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://jurnal.ugm.ac.id/ijc/article/viewFile/65951/31948",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "9411",
"original-title": [],
"page": "1252",
"prefix": "10.22146",
"published": {
"date-parts": [
[
2021,
8,
31
]
]
},
"published-online": {
"date-parts": [
[
2021,
8,
31
]
]
},
"publisher": "Universitas Gadjah Mada",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://jurnal.ugm.ac.id/ijc/article/view/65951"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Exploring the Potency of <i>Nigella sativa</i> Seed in Inhibiting SARS-CoV-2 Main Protease Using Molecular Docking and Molecular Dynamics Simulations",
"type": "journal-article",
"volume": "21"
}