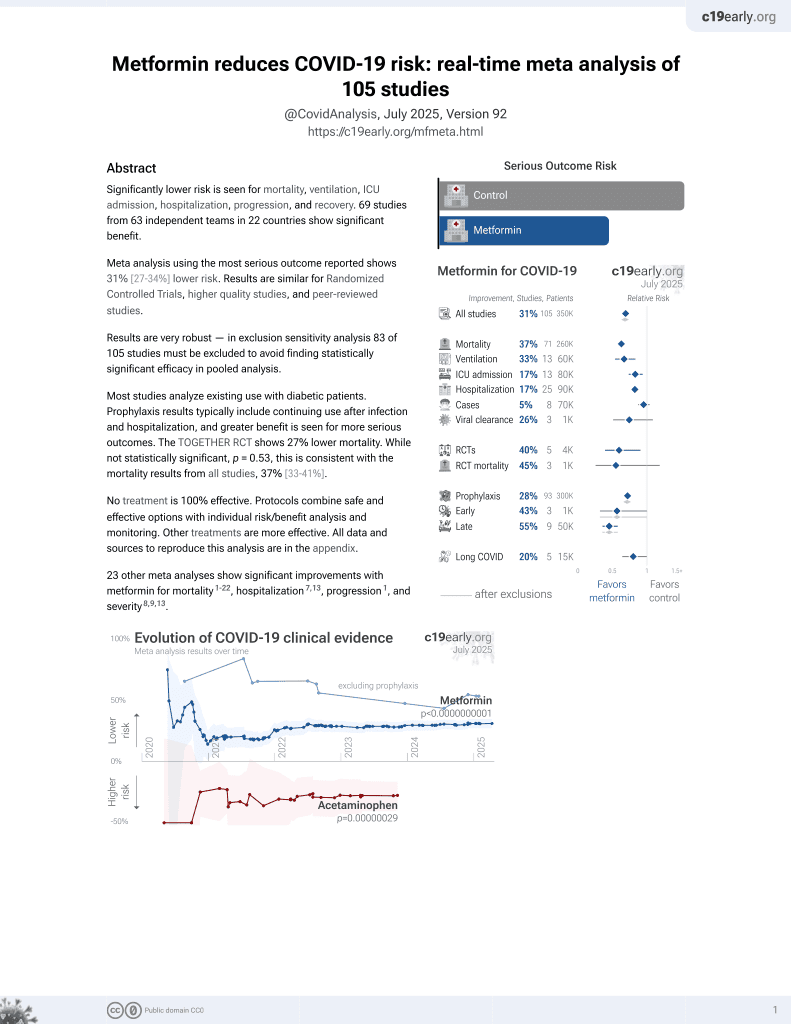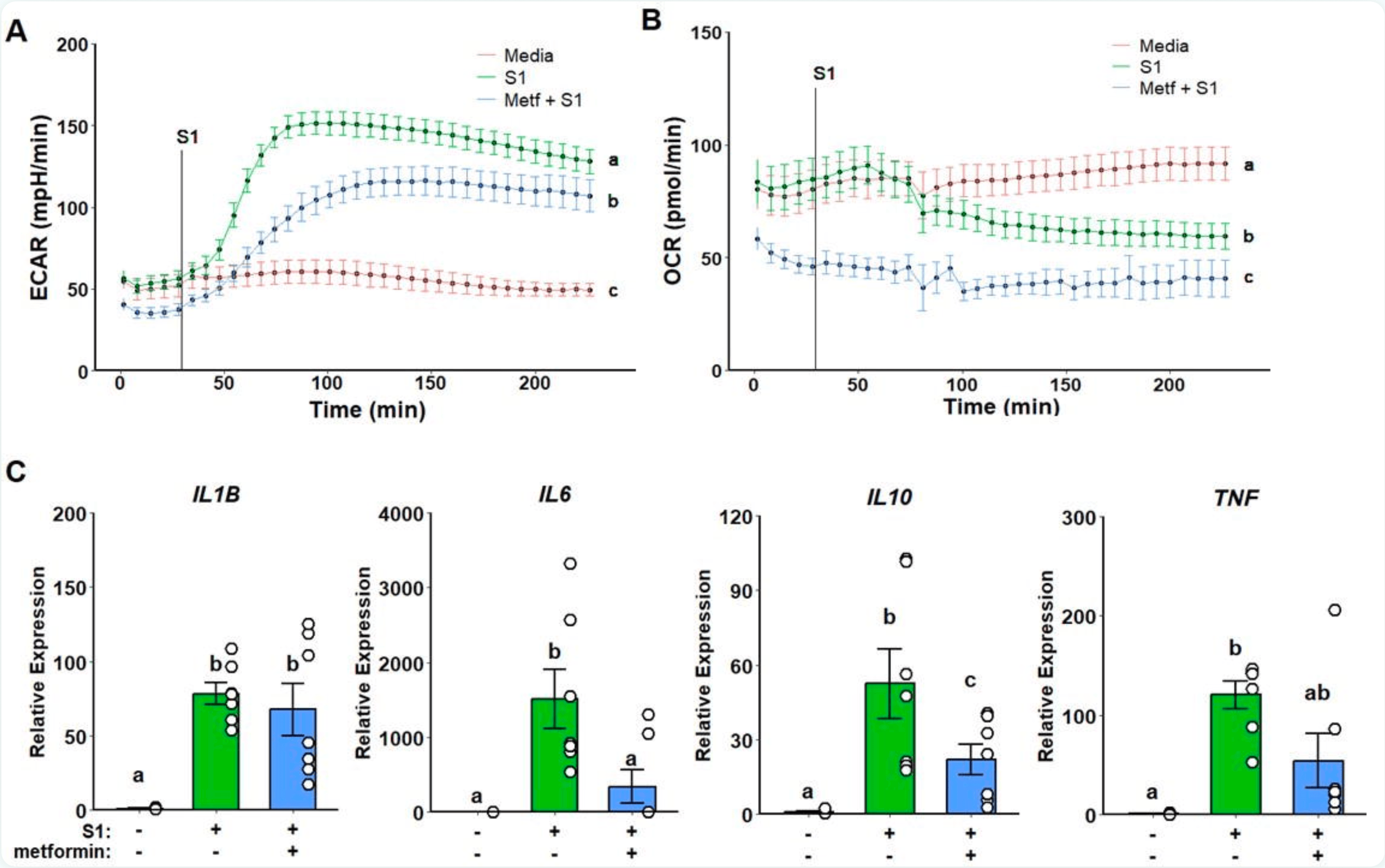
Immunoregulatory effect of metformin in monocytes exposed to SARS-CoV-2 spike protein subunit 1
et al., bioRxiv, doi:10.1101/2025.09.12.675877, Sep 2025
Metformin for COVID-19
3rd treatment shown to reduce risk in
July 2020, now with p < 0.00000000001 from 110 studies.
Lower risk for mortality, ventilation, ICU, hospitalization, progression, recovery, and viral clearance.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study showing that metformin suppresses inflammatory responses in human monocytes exposed to SARS-CoV-2 spike protein subunit 1.
18 preclinical studies support the efficacy of metformin for COVID-19:
A systematic review and meta-analysis of 15 non-COVID-19 preclinical studies showed that metformin inhibits pulmonary inflammation and oxidative stress, minimizes lung injury, and improves survival in animal models of acute respiratory distress syndrome (ARDS) or acute lung injury (ALI)15.
Metformin inhibits SARS-CoV-2 in vitro11,12, minimizes LPS-induced cytokine storm in a mouse model14, minimizes lung damage and fibrosis in a mouse model of LPS-induced ARDS10, may protect against SARS-CoV-2-induced neurological disorders9, may be beneficial via inhibitory effects on ORF3a-mediated inflammasome activation16, reduces UUO and FAN-induced kidney fibrosis10, increases mitochondrial function and decreases TGF-β-induced fibrosis, apoptosis, and inflammation markers in lung epithelial cells10, may reduce inflammation, oxidative stress, and thrombosis via regulating glucose metabolism2, attenuates spike protein S1-induced inflammatory response and α-synuclein aggregation8, may protect against COVID-19 cognitive impairment by suppressing HIF-1α stabilization and reducing neurodegenerative protein aggregation13, may reduce COVID-19 severity and long COVID by inhibiting NETosis via suppression of protein kinase C activation17, enhances interferon responses and reduces SARS-CoV-2 infection and inflammation in diabetic models by suppressing HIF-1α signaling7, may improve COVID-19 outcomes by preventing VDAC1 mistargeting to the plasma membrane, reducing ATP loss, and preserving immune cell function during cytokine storm18, reduces hyperglycemia-induced hepatic ACE2/TMPRSS2 up-regulation and SARS-CoV-2 entry6, may reduce COVID-19 severity by suppressing monocyte inflammatory responses and glycolytic activation via AMPK pathway modulation5, and may improve outcomes via modulation of immune responses with increased anti-inflammatory T lymphocyte gene expression and via enhanced gut microbiota diversity19.
1.
Tavares et al., Investigation of Interactions Between the Protein MPro and the Vanadium Complex VO(metf)2∙H2O: A Computational Approach for COVID-19 Treatment, Biophysica, doi:10.3390/biophysica5010004.
2.
Hou et al., Metformin is a potential therapeutic for COVID-19/LUAD by regulating glucose metabolism, Scientific Reports, doi:10.1038/s41598-024-63081-0.
3.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
4.
Lockwood, T., Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent, BioMetals, doi:10.1007/s10534-024-00590-5.
5.
Maurmann et al., Immunoregulatory effect of metformin in monocytes exposed to SARS-CoV-2 spike protein subunit 1, bioRxiv, doi:10.1101/2025.09.12.675877.
6.
Rao et al., Pathological Glucose Levels Enhance Entry Factor Expression and Hepatic SARS‐CoV‐2 Infection, Journal of Cellular and Molecular Medicine, doi:10.1111/jcmm.70581.
7.
Joshi et al., Severe SARS‐CoV‐2 infection in diabetes was rescued in mice supplemented with metformin and/or αKG, and patients taking metformin, via HIF1α‐IFN axis, Clinical and Translational Medicine, doi:10.1002/ctm2.70275.
8.
Chang et al., SARS-CoV-2 Spike Protein 1 Causes Aggregation of α-Synuclein via Microglia-Induced Inflammation and Production of Mitochondrial ROS: Potential Therapeutic Applications of Metformin, Biomedicines, doi:10.3390/biomedicines12061223.
9.
Yang et al., SARS-CoV-2 infection causes dopaminergic neuron senescence, Cell Stem Cell, doi:10.1016/j.stem.2023.12.012.
10.
Miguel et al., Enhanced fatty acid oxidation through metformin and baicalin as therapy for COVID-19 and associated inflammatory states in lung and kidney, Redox Biology, doi:10.1016/j.redox.2023.102957.
11.
Ventura-López et al., Treatment with metformin glycinate reduces SARS-CoV-2 viral load: An in vitro model and randomized, double-blind, Phase IIb clinical trial, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113223.
12.
Parthasarathy et al., Metformin Suppresses SARS-CoV-2 in Cell Culture, bioRxiv, doi:10.1101/2021.11.18.469078.
13.
Lee et al., SARS-CoV-2 spike protein causes synaptic dysfunction and p-tau and α-synuclein aggregation leading cognitive impairment: The protective role of metformin, PLOS One, doi:10.1371/journal.pone.0336015.
14.
Taher et al., Anti‑inflammatory effect of metformin against an experimental model of LPS‑induced cytokine storm, Experimental and Therapeutic Medicine, doi:10.3892/etm.2023.12114.
15.
Wang et al., Effects of metformin on acute respiratory distress syndrome in preclinical studies: a systematic review and meta-analysis, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1215307.
16.
Zhang et al., SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects, Pathogens, doi:10.3390/pathogens13010075.
17.
Monsalve et al., NETosis: A key player in autoimmunity, COVID-19, and long COVID, Journal of Translational Autoimmunity, doi:10.1016/j.jtauto.2025.100280.
Maurmann et al., 12 Sep 2025, USA, preprint, 5 authors.
Contact: bdpence@memphis.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Immunoregulatory effect of metformin in monocytes exposed to SARS-CoV-2 spike protein subunit 1
doi:10.1101/2025.09.12.675877
Background: Severe COVID-19 is characterized by a hyperinflammatory state associated with an exacerbated inflammatory activation of monocytes and macrophages in the respiratory tract. Metformin has been identified as a potent monocyte inflammatory suppressor, and it has been demonstrated to attenuate inflammation in COVID-19. The mechanisms underlying metformin anti-inflammatory effects are, however, unclear. We thus sought to investigate metformin's main interactions and their respective isolated effects in modulating monocyte inflammatory response to SARS-CoV-2 stimulation.
Methods : Classical human monocytes were isolated from healthy 18-40-year-old individuals and stimulated in vitro with recombinant spike protein subunit 1 (rS1) to assess glycolytic and oxidative metabolic responses by Seahorse extracellular flux analysis, and inflammatory gene expression by qPCR. Stimulated monocytes were either pre-treated with metformin, rotenone, S1QEL, or A769662. Results: Monocytes stimulated in vitro with rS1 showed an increased glycolytic response associated with production of pro-inflammatory cytokines. Metformin pre-treatment reduced glycolytic activation while partially suppressing inflammation. Rotenone-dependent mitochondrial complex I inhibition was not able to replicate the same effect, and neither complex I specific ROS scavenging. Conversely, A769662 induced AMPK activation led to suppressed glycolytic inflammatory response and cytokine expression pattern similar to metformin, thus suggesting AMPK modulation as a possible central component for metformin's mode of action upon S1 stimulation. Conclusions: In summary, further investigation into the interactions underlying AMPK activity on monocytes in the context of SARS-CoV-2 may provide a better elucidation of metformin's anti-inflammatory effect.
Author Contributions BP conceived the study. BP designed experiments. RMM, KD, NM, BLS, and BP collected data. RMM and BP analyzed data. RMM prepared the first manuscript draft. BP edited the manuscript draft. All authors read and approved the final manuscript.
References
Barek, Aziz, Islam, Impact of age, sex, comorbidities and clinical symptoms on the severity of COVID-19 cases: A meta-analysis with 55 studies and 10014 cases, Heliyon, doi:10.1016/j.heliyon.2020.e05684
Beigmohammadi, Jahanbin, Safaei, Pathological findings of postmortem biopsies from lung, heart, and liver of 7 deceased COVID-19 patients, Int J Surg Pathol, doi:10.1177/1066896920935195
Bułdak, Machnik, Bułdak, Łabuzek, Bołdys et al., Exenatide and metformin express their anti-inflammatory effects on human monocytes/macrophages by the attenuation of MAPKs and NFκB signaling, Naunyn Schmiedebergs Arch Pharmacol, doi:10.1007/s00210-016-1277-8
Chen, The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv, doi:10.1101/2020.03.27.20045427
Chen, Wu, Guo, Clinical and immunological features of severe and moderate coronavirus disease 2019, J Clin Invest, doi:10.1172/JCI137244
Cheng, Liu, Li, Metformin is associated with higher incidence of acidosis, but not mortality, in individuals with COVID-19 and pre-existing type 2 diabetes, Cell Metab, doi:10.1016/j.cmet.2020.08.013
Core, R: A language and environment for statistical computing
Cory, Emmons, Yarbro, Davis, Pence, Metformin suppresses monocyte immunometabolic activation by SARS-CoV-2 spike protein subunit 1, Front Immunol, doi:10.3389/fimmu.2021.733921
Foretz, Guigas, Bertrand, Pollak, Viollet, Metformin: From mechanisms of action to therapies, Cell Metab, doi:10.1016/j.cmet.2014.09.018
Hariyanto, Kurniawan, Metformin use is associated with reduced mortality rate from coronavirus disease 2019 (COVID-19) infection, Obes Med, doi:10.1016/j.obmed.2020.100290
Holm, A simple sequentially rejective multiple test procedure, Scand J Stat, doi:10.6084/m9.figshare.30113695
Jin, Cheng, Liu, Improvement of functional recovery by chronic metformin treatment is associated with enhanced alternative activation of microglia/macrophages and increased angiogenesis and neurogenesis following experimental stroke, Brain Behav Immun, doi:10.1016/j.bbi.2014.03.003
Jing, Wu, Li, Yang, Li et al., Metformin improves obesity-associated inflammation by altering macrophages polarization, Mol Cell Endocrinol, doi:10.1016/j.mce.2017.09.025
Karwaciak, Sałkowska, Karaś, Dastych, Ratajewski, Nucleocapsid and spike proteins of the coronavirus SARS-CoV-2 induce IL6 in monocytes and macrophagespotential implications for cytokine storm syndrome, Vaccines, doi:10.3390/vaccines9010054
Kelly, Tannahill, Murphy, Neill, Metformin inhibits the production of reactive oxygen species from NADH:ubiquinone oxidoreductase to limit induction of interleukin-1β (IL-1β) and boosts interleukin-10 (IL-10) in lipopolysaccharide (LPS)activated macrophages, J Biol Chem, doi:10.1074/jbc.M115.662114
Kim, Kwak, Cha, Metformin suppresses lipopolysaccharide (LPS)-induced inflammatory response in murine macrophages via activating transcription factor-3 (ATF-3) induction, J Biol Chem, doi:10.1074/jbc.M114.577908
Lai, Chen, Chao, Lee, Ko et al., COVID-19 vaccines: concerns beyond protective efficacy and safety, Expert Rev Vaccines, doi:10.1080/14760584.2021.1949293
Laing, Lorenc, Molino, Barrio, A dynamic COVID-19 immune signature includes associations with poor prognosis, Nat Med, doi:10.1038/s41591-020-1038-6
Li, Ragheb, Lawler, Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production, J Biol Chem, doi:10.1074/jbc.M210432200
Li, Xu, Mihaylova, AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice, Cell Metab, doi:10.1016/j.cmet.2011.03.009
Liao, Liu, Yuan, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat Med, doi:10.1038/s41591-020-0901-9
Livak, Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods, doi:10.1006/meth.2001.1262
Lukito, Pranata, Henrina, Lim, Lawrensia et al., The effect of metformin consumption on mortality in hospitalized COVID-19 patients: A systematic review and meta-analysis, Diabetes Metab Syndr, doi:10.1016/j.dsx.2020.11.006
Mann, Menon, Knight, Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19, Sci Immunol, doi:10.1126/sciimmunol.abd6197
Markov, Ghafari, Beer, The evolution of SARS-CoV-2, Nat Rev Microbiol, doi:10.1038/s41579-023-00878-2
Mehta, Mcauley, Brown, COVID-19: Consider cytokine storm syndromes and immunosuppression, Lancet, doi:10.1016/S0140-6736(20)30628-0
O'neill, Hardie, Metabolism of inflammation limited by AMPK and pseudostarvation, Nature, doi:10.1038/nature11862
Pan, Sze, Minhas, The impact of ethnicity on clinical outcomes in COVID-19: A systematic review, EClinicalMedicine, doi:10.1016/j.eclinm.2020.100404
Pence, Yarbro, Aging impairs mitochondrial respiratory capacity in classical monocytes, Exp Gerontol, doi:10.1016/j.exger.2018.04.008
Qing, Fu, Wu, Zhou, Yu et al., Metformin induces the M2 macrophage polarization to accelerate the wound healing via regulating AMPK/mTOR/NLRP3 inflammasome singling pathway, Am J Transl Res
Ramachandran, Dobie, Jr, Resolving the fibrotic niche of human liver cirrhosis at single-cell level, Nature, doi:10.1038/s41586-019-1631-3
Ropa, Cooper, Capitano, Van't Hof, Broxmeyer, Human hematopoietic stem, progenitor, and immune cells respond ex vivo to SARS-CoV-2 spike protein, Stem Cell Rev Rep, doi:10.1007/s12015-020-10056-z
Schulert, Grom, Pathogenesis of macrophage activation syndrome and potential for cytokine-directed therapies, Annu Rev Med, doi:10.1146/annurev-med-061813-012806
Shaheen, Sambas, Alenezi, Alharbi, Aldibasi et al., COVID-19 reinfection: A multicenter retrospective study in Saudi Arabia, Ann Thorac Med, doi:10.4103/atm.atm_74_22
Soberanes, Misharin, Jairaman, Metformin targets mitochondrial electron transport to reduce air-pollution-induced thrombosis, Cell Metab, doi:10.1016/j.cmet.2018.09.019
Toussi, Hammond, Gerstenberger, Anderson, Therapeutics for COVID-19, Nat Microbiol, doi:10.1038/s41564-023-01356-4
Ursini, Russo, Pellino, Metformin and autoimmunity: A "new deal" of an old drug, Front Immunol, doi:10.3389/fimmu.2018.01236
Valle, Kim-Schulze, Huang, An inflammatory cytokine signature predicts COVID-19 severity and survival, Nat Med, doi:10.1038/s41591-020-1051-9
Vasamsetti, Karnewar, Kanugula, Thatipalli, Kumar et al., Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: Potential role in atherosclerosis, Diabetes, doi:10.2337/db14-1225
Wang, Li, Wang, Metformin inhibits metastatic breast cancer progression and improves chemosensitivity by inducing vessel normalization via PDGF-B downregulation, J Exp Clin Cancer Res, doi:10.1186/s13046-019-1211-2
Wen, Su, Tang, Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing, Cell Discov, doi:10.1038/s41421-020-00187-5
Xian, Liu, Nilsson, Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation, Immunity, doi:10.1016/j.immuni.2021.05.004
Yu, Zhu, Huang, Metformin relieves acute respiratory distress syndrome by reducing miR-138 expression, Eur Rev Med Pharmacol Sci, doi:10.26355/eurrev_201808_15737
Zhang, Guo, Lei, Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes, J Leukoc Biol, doi:10.1002/JLB.4HI0720-470R
Zhou, Ren, Zhang, Heightened innate immune responses in the respiratory tract of COVID-19 patients, Cell Host Microbe, doi:10.1016/j.chom.2020.04.017
DOI record:
{
"DOI": "10.1101/2025.09.12.675877",
"URL": "http://dx.doi.org/10.1101/2025.09.12.675877",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>Severe COVID-19 is characterized by a hyperinflammatory state associated with an exacerbated inflammatory activation of monocytes and macrophages in the respiratory tract. Metformin has been identified as a potent monocyte inflammatory suppressor, and it has been demonstrated to attenuate inflammation in COVID-19. The mechanisms underlying metformin anti-inflammatory effects are, however, unclear. We thus sought to investigate metformin’s main interactions and their respective isolated effects in modulating monocyte inflammatory response to SARS-CoV-2 stimulation.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>Classical human monocytes were isolated from healthy 18-40-year-old individuals and stimulated <jats:italic>in vitro</jats:italic> with recombinant spike protein subunit 1 (rS1) to assess glycolytic and oxidative metabolic responses by Seahorse extracellular flux analysis, and inflammatory gene expression by qPCR. Stimulated monocytes were either pre-treated with metformin, rotenone, S1QEL, or A769662.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>Monocytes stimulated <jats:italic>in vitro</jats:italic> with rS1 showed an increased glycolytic response associated with production of pro-inflammatory cytokines. Metformin pre-treatment reduced glycolytic activation while partially suppressing inflammation. Rotenone-dependent mitochondrial complex I inhibition was not able to replicate the same effect, and neither complex I specific ROS scavenging. Conversely, A769662 induced AMPK activation led to suppressed glycolytic inflammatory response and cytokine expression pattern similar to metformin, thus suggesting AMPK modulation as a possible central component for metformin’s mode of action upon S1 stimulation.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Conclusions</jats:title>\n <jats:p>In summary, further investigation into the interactions underlying AMPK activity on monocytes in the context of SARS-CoV-2 may provide a better elucidation of metformin’s anti-inflammatory effect.</jats:p>\n </jats:sec>",
"accepted": {
"date-parts": [
[
2025,
9,
12
]
]
},
"author": [
{
"ORCID": "https://orcid.org/0000-0003-1759-1778",
"affiliation": [],
"authenticated-orcid": false,
"family": "Maurmann",
"given": "Rafael Moura",
"sequence": "first"
},
{
"affiliation": [],
"family": "Davis",
"given": "Kierstin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7518-8066",
"affiliation": [],
"authenticated-orcid": false,
"family": "Mosalmanzadeh",
"given": "Negin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5823-2611",
"affiliation": [],
"authenticated-orcid": false,
"family": "Schmitt",
"given": "Brenda Landvoigt",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4059-9092",
"affiliation": [],
"authenticated-orcid": false,
"family": "Pence",
"given": "Brandt D.",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
9,
13
]
],
"date-time": "2025-09-13T03:55:13Z",
"timestamp": 1757735713000
},
"deposited": {
"date-parts": [
[
2025,
9,
16
]
],
"date-time": "2025-09-16T18:35:19Z",
"timestamp": 1758047719000
},
"funder": [
{
"name": "University of Memphis / University of Tennessee Health Science Center CORNET"
},
{
"DOI": "10.13039/100000968",
"award": [
"18AIREA33961089"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100000968",
"id-type": "DOI"
}
],
"name": "American Heart Association"
},
{
"DOI": "10.13039/100000968",
"award": [
"19TPA34910232"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100000968",
"id-type": "DOI"
}
],
"name": "American Heart Association"
},
{
"name": "University of Memphis College of Health Sciences"
}
],
"group-title": "Immunology",
"indexed": {
"date-parts": [
[
2025,
9,
18
]
],
"date-time": "2025-09-18T16:21:41Z",
"timestamp": 1758212501246,
"version": "3.44.0"
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
9,
12
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
12
]
],
"date-time": "2025-09-12T00:00:00Z",
"timestamp": 1757635200000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2025.09.12.675877",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2025,
9,
12
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2025,
9,
12
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"key": "2025091611351074000_2025.09.12.675877v1.1",
"unstructured": "World Health Organization. WHO COVID-19 dashboard. World Health Organization. Updated February, 2025. Accessed February 11, 2025. https://covid19.who.int/"
},
{
"DOI": "10.1080/14760584.2021.1949293",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.2"
},
{
"DOI": "10.1038/s41579-023-00878-2",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.3"
},
{
"DOI": "10.1016/j.heliyon.2020.e05684",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.4"
},
{
"DOI": "10.4103/atm.atm_74_22",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.5"
},
{
"DOI": "10.1172/JCI137244",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.6"
},
{
"DOI": "10.1038/s41591-020-1038-6",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.7"
},
{
"DOI": "10.1038/s41591-020-1051-9",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.8"
},
{
"DOI": "10.1016/j.eclinm.2020.100404",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.9"
},
{
"DOI": "10.1016/S0140-6736(20)30628-0",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.10"
},
{
"DOI": "10.1146/annurev-med-061813-012806",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.11"
},
{
"DOI": "10.1038/s41591-020-0901-9",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.12"
},
{
"DOI": "10.1016/j.chom.2020.04.017",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.13"
},
{
"DOI": "10.1101/2020.03.27.20045427",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.14"
},
{
"DOI": "10.1177/1066896920935195",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.15"
},
{
"DOI": "10.1038/s41586-019-1631-3",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.16"
},
{
"DOI": "10.1002/JLB.4HI0720-470R",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.17"
},
{
"DOI": "10.1038/s41421-020-00187-5",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.18"
},
{
"DOI": "10.1126/sciimmunol.abd6197",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.19"
},
{
"DOI": "10.1038/s41564-023-01356-4",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.20"
},
{
"DOI": "10.1016/j.obmed.2020.100290",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.21"
},
{
"DOI": "10.1016/j.dsx.2020.11.006",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.22"
},
{
"DOI": "10.1016/j.cmet.2020.08.013",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.23"
},
{
"DOI": "10.1016/j.immuni.2021.05.004",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.24"
},
{
"DOI": "10.26355/eurrev_201808_15737",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.25"
},
{
"DOI": "10.1016/j.cmet.2018.09.019",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.26"
},
{
"DOI": "10.1074/jbc.M115.662114",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.27"
},
{
"DOI": "10.2337/db14-1225",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.28"
},
{
"DOI": "10.1074/jbc.M114.577908",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.29"
},
{
"DOI": "10.1016/j.mce.2017.09.025",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.30"
},
{
"DOI": "10.1007/s00210-016-1277-8",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.31"
},
{
"DOI": "10.3389/fimmu.2018.01236",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.32"
},
{
"DOI": "10.1016/j.cmet.2014.09.018",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.33"
},
{
"DOI": "10.3389/fimmu.2021.733921",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.34"
},
{
"DOI": "10.1016/j.exger.2018.04.008",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.35"
},
{
"DOI": "10.1006/meth.2001.1262",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.36"
},
{
"key": "2025091611351074000_2025.09.12.675877v1.37",
"unstructured": "R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2014. Available from: https://www.R-project.org/"
},
{
"article-title": "A simple sequentially rejective multiple test procedure",
"first-page": "65",
"issue": "2",
"journal-title": "Scand J Stat",
"key": "2025091611351074000_2025.09.12.675877v1.38",
"volume": "6",
"year": "1979"
},
{
"DOI": "10.6084/m9.figshare.30113695",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.39"
},
{
"DOI": "10.1007/s12015-020-10056-z",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.40"
},
{
"DOI": "10.3390/vaccines9010054",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.41"
},
{
"DOI": "10.1074/jbc.M210432200",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.42"
},
{
"DOI": "10.1016/j.cmet.2011.03.009",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.43"
},
{
"DOI": "10.1038/nature11862",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.44"
},
{
"DOI": "10.1016/j.bbi.2014.03.003",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.45"
},
{
"article-title": "Metformin induces the M2 macrophage polarization to accelerate the wound healing via regulating AMPK/mTOR/NLRP3 inflammasome singling pathway",
"first-page": "655",
"issue": "2",
"journal-title": "Am J Transl Res",
"key": "2025091611351074000_2025.09.12.675877v1.46",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1186/s13046-019-1211-2",
"doi-asserted-by": "publisher",
"key": "2025091611351074000_2025.09.12.675877v1.47"
}
],
"reference-count": 47,
"references-count": 47,
"relation": {},
"resource": {
"primary": {
"URL": "http://biorxiv.org/lookup/doi/10.1101/2025.09.12.675877"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Immunoregulatory effect of metformin in monocytes exposed to SARS-CoV-2 spike protein subunit 1",
"type": "posted-content"
}