SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects
et al., Pathogens, doi:10.3390/pathogens13010075, Jan 2024
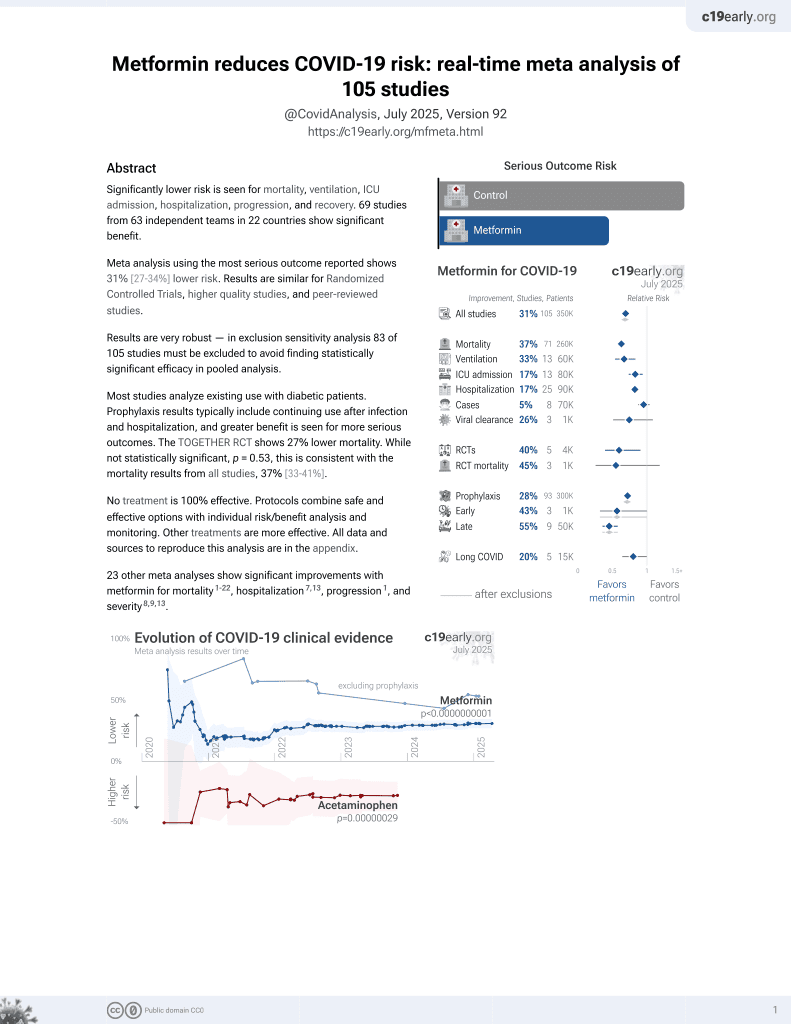
Metformin for COVID-19
3rd treatment shown to reduce risk in
July 2020, now with p < 0.00000000001 from 110 studies.
Lower risk for mortality, ventilation, ICU, hospitalization, progression, recovery, and viral clearance.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Review of the ORF3a protein of SARS-CoV-2 as a therapeutic target for COVID-19 due to its role in viral pathogenesis, triggering cytokine storms, tissue damage, and disease severity. Existing drugs doxycycline, metformin, and ibrutinib are proposed for inhibiting ORF3a-mediated pathways, and authors identify chlorin e6 and TmPyP4 as promising inhibitors.
1.
Monsalve et al., NETosis: A key player in autoimmunity, COVID-19, and long COVID, Journal of Translational Autoimmunity, doi:10.1016/j.jtauto.2025.100280.
2.
Halabitska et al., Metformin in Antiviral Therapy: Evidence and Perspectives, Viruses, doi:10.3390/v16121938.
3.
Plowman et al., Anti-Inflammatory Potential of the Anti-Diabetic Drug Metformin in the Prevention of Inflammatory Complications and Infectious Diseases Including COVID-19: A Narrative Review, International Journal of Molecular Sciences, doi:10.3390/ijms25105190.
4.
De Jesús-González et al., A Dual Pharmacological Strategy against COVID-19: The Therapeutic Potential of Metformin and Atorvastatin, Microorganisms, doi:10.3390/microorganisms12020383.
5.
Halma et al., Exploring autophagy in treating SARS-CoV-2 spike protein-related pathology, Endocrine and Metabolic Science, doi:10.1016/j.endmts.2024.100163.
6.
Zhang et al., SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects, Pathogens, doi:10.3390/pathogens13010075.
7.
Gomaa et al., Pharmacological evaluation of vitamin D in COVID-19 and long COVID-19: recent studies confirm clinical validation and highlight metformin to improve VDR sensitivity and efficacy, Inflammopharmacology, doi:10.1007/s10787-023-01383-x.
Zhang et al., 14 Jan 2024, USA, peer-reviewed, 10 authors.
Contact: rzhao@som.umaryland.edu (corresponding author), jiantao.zhang@som.umaryland.edu, chenyu.zhang@som.umaryland.edu, hom@rx.umaryland.edu, fxue@rx.umaryland.edu, mnasr@niaid.nih.gov, vgerzanich@som.umaryland.edu, msimard@som.umaryland.edu, zhangyj@umd.edu, qiyi.tang@howard.edu.
SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects
Pathogens, doi:10.3390/pathogens13010075
The COVID-19 pandemic caused by SARS-CoV-2 has posed unparalleled challenges due to its rapid transmission, ability to mutate, high mortality and morbidity, and enduring health complications. Vaccines have exhibited effectiveness, but their efficacy diminishes over time while new variants continue to emerge. Antiviral medications offer a viable alternative, but their success has been inconsistent. Therefore, there remains an ongoing need to identify innovative antiviral drugs for treating COVID-19 and its post-infection complications. The ORF3a (open reading frame 3a) protein found in SARS-CoV-2, represents a promising target for antiviral treatment due to its multifaceted role in viral pathogenesis, cytokine storms, disease severity, and mortality. ORF3a contributes significantly to viral pathogenesis by facilitating viral assembly and release, essential processes in the viral life cycle, while also suppressing the body's antiviral responses, thus aiding viral replication. ORF3a also has been implicated in triggering excessive inflammation, characterized by NF-κB-mediated cytokine production, ultimately leading to apoptotic cell death and tissue damage in the lungs, kidneys, and the central nervous system. Additionally, ORF3a triggers the activation of the NLRP3 inflammasome, inciting a cytokine storm, which is a major contributor to the severity of the disease and subsequent mortality. As with the spike protein, ORF3a also undergoes mutations, and certain mutant variants correlate with heightened disease severity in COVID-19. These mutations may influence viral replication and host cellular inflammatory responses. While establishing a direct link between ORF3a and mortality is difficult, its involvement in promoting inflammation and exacerbating disease severity likely contributes to higher mortality rates in severe COVID-19 cases. This review offers a comprehensive and detailed exploration of ORF3a's potential as an innovative antiviral drug target. Additionally, we outline potential strategies for discovering and developing ORF3a inhibitor drugs to counteract its harmful effects, alleviate tissue damage, and reduce the severity of COVID-19 and its lingering complications.
Author Contributions: Conceptualization, R.Y.Z.; writing-original draft preparation, R.Y.Z. and J.Z.; writing-review and editing, J.M.S., F.X., Q.T., Y.Z., V.G., M.N., K.H. and C.Z.; visualization, J.Z.; supervision, R.Y.Z.; project administration, R.Y.Z.; funding acquisition, R.Y.Z., J.M.S., Q.T. and V.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement: Not applicable. Informed Consent Statement: Not applicable.
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
Akerstrom, Mirazimi, Tan, Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and, S. Antivir. Res, doi:10.1016/j.antiviral.2006.10.008
Alcendor, Matthews-Juarez, Smoot, Hildreth, Lamar et al., Breakthrough COVID-19 Infections in the US: Implications for Prolonging the Pandemic, Vaccines, doi:10.3390/vaccines10050755
Alexander, Mangalaparthi, Madugundu, Moyer, Adam et al., Acute Kidney Injury in Severe COVID-19 Has Similarities to Sepsis-Associated Kidney Injury: A Multi-Omics Study, Mayo Clin. Proc, doi:10.1016/j.mayocp.2021.07.001
Amraei, Yin, Napoleon, Suder, Berrigan et al., CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2, ACS Cent. Sci, doi:10.1021/acscentsci.0c01537
Andreola, Litvak, Yeast and the AIDS virus: The odd couple, J. Biomed. Biotechnol, doi:10.1155/2012/549020
Aranda-Rivera, Cruz-Gregorio, Aparicio-Trejo, Pedraza-Chaverri, Mitochondrial Redox Signaling and Oxidative Stress in Kidney Diseases, Biomolecules, doi:10.3390/biom11081144
Armaly, Kinaneh, Skorecki, Renal Manifestations of COVID-19: Physiology and Pathophysiology, J. Clin. Med, doi:10.3390/jcm10061216
Arora, Maiti, Differential biophysical behavior of human telomeric RNA and DNA quadruplex, J. Phys. Chem. B, doi:10.1021/jp810638n
Bakhshi, Shamsi, MCC950 in the treatment of NLRP3-mediated inflammatory diseases: Latest evidence and therapeutic outcomes, Int. Immunopharmacol, doi:10.1016/j.intimp.2022.108595
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the Treatment of COVID-19-Final Report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Benko, Elder, Li, Liang, Zhao, HIV-1 Protease in the Fission Yeast Schizosaccharomyces pombe, PLoS ONE, doi:10.1371/journal.pone.0151286
Benko, Elder, Liang, Zhao, Fission yeast as a HTS platform for molecular probes of HIV-1 Vpr-induced cell death, Int. J. High Throughput Screen
Benko, Liang, Li, Elder, Sarkar et al., A fission yeast cell-based system for multidrug resistant HIV-1 proteases, Cell Biosci, doi:10.1186/s13578-016-0131-5
Benko, Zhang, Zhao, Development of A Fission Yeast Cell-Based Platform for High Throughput Screening of HIV-1 Protease Inhibitors, Curr. HIV Res, doi:10.2174/1570162X17666191128102839
Bertoni, Penco, Mollica, Bocca, Prigione et al., Spontaneous NLRP3 inflammasome-driven IL1-beta secretion is induced in severe COVID-19 patients and responds to anakinra treatment, J. Allergy Clin. Immunol, doi:10.1016/j.jaci.2022.05.029
Bianchi, Borsetti, Ciccozzi, Pascarella, SARS-CoV-2 ORF3a: Mutability and function, Int. J. Biol. Macromol, doi:10.1016/j.ijbiomac.2020.12.142
Bolay, Karadas, Ozturk, Sonkaya, Tasdelen et al., HMGB1, NLRP3, IL-6 and ACE2 levels are elevated in COVID-19 with headache: A window to the infection-related headache mechanism, J. Headache Pain, doi:10.1186/s10194-021-01306-7
Bowe, Cai, Xie, Gibson, Maddukuri et al., Acute Kidney Injury in a National Cohort of Hospitalized US Veterans with COVID-19, Clin. J. Am. Soc. Nephrol, doi:10.2215/CJN.09610620
Brant, Tian, Majerciak, Yang, Zheng, SARS-CoV-2: From its discovery to genome structure, transcription, and replication, Cell Biosci, doi:10.1186/s13578-021-00643-z
Braun, Lutgehetmann, Pfefferle, Wong, Carsten et al., SARS-CoV-2 renal tropism associates with acute kidney injury, Lancet, doi:10.1016/S0140-6736(20)31759-1
Breitinger, Farag, Sticht, Breitinger, Viroporins, Structure, function, and their role in the life cycle of SARS-CoV-2, Int. J. Biochem. Cell Biol, doi:10.1016/j.biocel.2022.106185
Busscher, Befekadu, Liu, Xiao, SARS-CoV-2 ORF3a-Mediated NF-kappaB Activation Is Not Dependent on TRAF-Binding Sequence, Viruses, doi:10.3390/v15112229
Cai, Chen, Feng, Asadi, Kaufman et al., SARS-CoV-2 viral protein ORF3A injures renal tubules by interacting with TRIM59 to induce STAT3 activation, Mol. Ther, doi:10.1016/j.ymthe.2022.12.008
Caillet-Saguy, Durbesson, Rezelj, Gogl, Tran et al., Host PDZ-containing proteins targeted by SARS-CoV-2, FEBS J, doi:10.1111/febs.15881
Camerini, Randall, Trappl-Kimmons, Oberai, Hung et al., Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray, Microbiol. Spectr, doi:10.1128/Spectrum.01416-21
Castano-Rodriguez, Honrubia, Gutierrez-Alvarez, Dediego, Nieto-Torres et al., Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis, mBio, doi:10.1128/mBio.02325-17
Chan, Chaudhary, Saha, Chauhan, Vaid et al., AKI in Hospitalized Patients with COVID-19, J. Am. Soc. Nephrol, doi:10.1681/ASN.2020050615
Chen, Long, Xu, Tan, Wang et al., Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients, Cell Mol. Immunol, doi:10.1038/s41423-020-0492-x
Chen, Moriyama, Chang, Ichinohe, Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome, Front. Microbiol, doi:10.3389/fmicb.2019.00050
Chen, Zheng, Sun, Ji, Li et al., ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress, Dev. Cell, doi:10.1016/j.devcel.2021.10.006
Cheng, Zhang, Chen, Wang, Zhang et al., Dynamic landscape mapping of humoral immunity to SARS-CoV-2 identifies non-structural protein antibodies associated with the survival of critical COVID-19 patients, Signal Transduct. Target. Ther, doi:10.1038/s41392-021-00718-w
Chu, Chan, Yuen, Shuai, Yuan et al., Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study, Lancet Microbe, doi:10.1016/S2666-5247(20)30004-5
Copur, Berkkan, Basile, Tuttle, Kanbay, Post-acute COVID-19 syndrome and kidney diseases: What do we know?, J. Nephrol, doi:10.1007/s40620-022-01296-y
Crunfli, Carregari, Veras, Silva, Nogueira et al., Morphological, cellular, and molecular basis of brain infection in COVID-19 patients, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2200960119
Cruz-Cosme, Zhang, Liu, Mahase, Sallapalli et al., A novel diG motif in ORF3a protein of SARS-CoV-2 for intracellular transport, Front. Cell Dev. Biol
De Silva, Liu, Lindsey, Dong, Moore et al., The impact of viral mutations on recognition by SARS-CoV-2 specific T-cells, iScience, doi:10.1038/d41586-022-00057-y
Dedoni, Avdoshina, Camoglio, Siddi, Fratta et al., K18-and CAG-hACE2 Transgenic Mouse Models and SARS-CoV-2: Implications for Neurodegeneration Research, Molecules, doi:10.3390/molecules27134142
Diao, Wang, Wang, Feng, Zhang et al., Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection, Nat. Commun, doi:10.1038/s41467-021-22781-1
Dong, Mead, Tian, Park, Garcia et al., The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus, J. Virol, doi:10.1128/JVI.00964-21
Donia, Bokhari, Apoptosis induced by SARS-CoV-2: Can we target it?, Apoptosis, doi:10.1007/s10495-021-01656-2
Duarte, Vazquez, Diethelm-Varela, Pavez, Berrios-Rojas et al., Differential Severe Acute Respiratory Syndrome Coronavirus 2-Specific Humoral Response in Inactivated Virus-Vaccinated, Convalescent, and Breakthrough-Infected Subjects, J. Infect. Dis, doi:10.1093/infdis/jiad320
Fam, Sedky, Turky, Breitinger, Breitinger, Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites, Sci. Rep, doi:10.1038/s41598-023-31764-9
Freeman, Swartz, Targeting the NLRP3 Inflammasome in Severe COVID-19, Front. Immunol, doi:10.3389/fimmu.2020.01518
Geri, Darmon, Zafrani, Fartoukh, Voiriot et al., Acute kidney injury in SARS-CoV2-related pneumonia ICU patients: A retrospective multicenter study, Ann. Intensive Care, doi:10.1186/s13613-021-00875-9
Ghosh, Dellibovi-Ragheb, Kerviel, Pak, Qiu et al., beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway, Cell, doi:10.1016/j.cell.2020.10.039
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Gowda, Patrick, Joshi, Kumawat, Sen, Glycyrrhizin prevents SARS-CoV-2 S1 and Orf3a induced high mobility group box 1 (HMGB1) release and inhibits viral replication, Cytokine, doi:10.1016/j.cyto.2021.155496
Grifoni, Weiskopf, Ramirez, Mateus, Dan et al., Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals, Cell, doi:10.1016/j.cell.2020.05.015
Group, Horby, Lim, Emberson, Mafham et al., Dexamethasone in Hospitalized Patients with COVID-19, N. Engl. J. Med, doi:10.1056/NEJMoa2021436
Guarnieri, Angelin, Murdock, Schaefer, Portluri et al., SARS-CoV-2 viroporins activate the NLRP3-inflammasome by the mitochondrial permeability transition pore, Front. Immunol, doi:10.3389/fimmu.2023.1064293
Guo, Liu, Yang, Liu, Xu et al., Effect of Thymoquinone on Acute Kidney Injury Induced by Sepsis in BALB/c Mice, Biomed. Res. Int, doi:10.1155/2020/1594726
Hachim, Kavian, Cohen, Chin, Chu et al., ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection, Nat. Immunol, doi:10.1038/s41590-020-0773-7
Han, Bailly, Abichandani, Thadhani, Bonventre, Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury, Kidney Int, doi:10.1046/j.1523-1755.2002.00433.x
Harcourt, Tamin, Lu, Kamili, Sakthivel et al., Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States, Emerg. Infect. Dis, doi:10.3201/eid2606.200516
He, Chang, Peng, Zhu, Liu et al., Glibenclamide Directly Prevents Neuroinflammation by Targeting SUR1-TRPM4-Mediated NLRP3 Inflammasome Activation in Microglia, Mol. Neurobiol, doi:10.1007/s12035-022-02998-x
Hoffmann, Kleine-Weber, Schroeder, Kruger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Honrubia, Gutierrez-Alvarez, Sanz-Bravo, Gonzalez-Miranda, Munoz-Santos et al., SARS-CoV-2-Mediated Lung Edema and Replication Are Diminished by Cystic Fibrosis Transmembrane Conductance Regulator Modulators, mBio, doi:10.1128/mbio.03136-22
Hu, Huang, Yin, The cytokine storm and COVID-19, J. Med. Virol, doi:10.1002/jmv.26232
Issa, Merhi, Panossian, Salloum, Tokajian, SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis, mSystems, doi:10.1128/mSystems.00266-20
Izadi, Brenner, Mahil, Dand, Yiu et al., Association Between Tumor Necrosis Factor Inhibitors and the Risk of Hospitalization or Death Among Patients with Immune-Mediated Inflammatory Disease and COVID-19, JAMA Netw. Open, doi:10.1001/jamanetworkopen.2021.29639
Jin, Du, Xu, Deng, Liu et al., Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Jin, Zheng, Tang, Mechanism of inflammasome activation by SARS coronavirus 3a protein: Abridged secondary publication, Hong Kong Med. J
Jorgensen, Miao, Pyroptotic cell death defends against intracellular pathogens, Immunol. Rev, doi:10.1111/imr.12287
Kakkanas, Karamichali, Koufogeorgou, Kotsakis, Georgopoulou et al., Targeting the YXXPhi Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions, Biomolecules, doi:10.3390/biom12081052
Kalejaiye, Bhattacharya, Burt, Travieso, Okafor et al., SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes, Front. Cell Dev. Biol, doi:10.3389/fcell.2022.855340
Kayagaki, Stowe, Lee, O'rourke, Anderson et al., Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling, Nature, doi:10.1038/nature15541
Kern, Sorum, Mali, Hoel, Sridharan et al., Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00619-0
Khan, Chen, Yang, Raghuram, Khundmiri et al., Does SARS-CoV-2 Infect the Kidney?, J. Am. Soc. Nephrol, doi:10.1681/ASN.2020081229
Laplantine, Chable-Bessia, Oudin, Swain, Soria et al., The FDA-approved drug Auranofin has a dual inhibitory effect on SARS-CoV-2 entry and NF-kappaB signaling, iScience, doi:10.1016/j.isci.2022.105066
Lebedeva, Gubarev, Mamardashvili, Zaitceva, Zdanovich et al., Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2, Sci. Rep, doi:10.1038/s41598-021-99072-8
Lednicky, Cherabuddi, Tagliamonte, Elbadry, Subramaniam et al., In-Frame 12-Nucleotide Deletion within Open Reading Frame 3a in a SARS-CoV-2 Strain Isolated from a Patient Hospitalized with COVID-19, Microbiol. Resour. Announc, doi:10.1128/MRA.00137-21
Lee, Jung, Kim, Charles, Christ et al., Super-resolution proximity labeling reveals anti-viral protein network and its structural changes against SARS-CoV-2 viral proteins, Cell Rep, doi:10.1016/j.celrep.2023.112835
Legrand, Bell, Forni, Joannidis, Koyner et al., Pathophysiology of COVID-19-associated acute kidney injury, Nat. Rev. Nephrol, doi:10.1038/s41581-021-00452-0
Li, Poulsen, Fenyvuesvolgyi, Yashiroda, Yoshida et al., Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1619735114
Li, Zhao, Molecular Cloning and Characterization of Small Viral Genome in Fission Yeast, Methods Mol. Biol, doi:10.1007/978-1-4939-7546-4_5
Liddelow, Guttenplan, Clarke, Bennett, Bohlen et al., Neurotoxic reactive astrocytes are induced by activated microglia, Nature, doi:10.1038/nature21029
Liu, Hsueh, Lin, Chiu, Kao et al., Disease-specific B Cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens, J. Infect. Dis, doi:10.1086/422753
Liu, Zhang, Liu, Xia, Zou et al., A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions, Nat. Commun, doi:10.1038/s41467-022-31930-z
Lonze, Spiegler, Wesson, Alachkar, Petkova et al., A Randomized Double-Blinded Placebo Controlled Trial of Clazakizumab for the Treatment of COVID-19 Pneumonia with Hyperinflammation, Crit. Care Med, doi:10.1097/CCM.0000000000005591
Lu, Zheng, Xu, Schwarz, Du et al., Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0605402103
Ma, Chen, Xue, Wang, Dong et al., Cinnamaldehyde inhibits cytokine storms induced by the ORF3a protein of SARS-CoV-2 via ROS-elimination in activated T cells, Phytother. Res, doi:10.1002/ptr.8016
Majumdar, Niyogi, ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection, Epidemiol. Infect, doi:10.1017/S0950268820002599
Marshall, How COVID-19 can damage the brain, Nature, doi:10.1038/d41586-020-02599-5
Martin-Sancho, Lewinski, Pache, Stoneham, Yin et al., Functional landscape of SARS-CoV-2 cellular restriction, Mol. Cell, doi:10.1016/j.molcel.2021.04.008
May, Cassol, Hannoudi, Larsen, Lerma et al., A multi-center retrospective cohort study defines the spectrum of kidney pathology in Coronavirus 2019 Disease (COVID-19), Kidney Int, doi:10.1016/j.kint.2021.07.015
Mcclenaghan, Hanson, Lee, Nichols, Coronavirus Proteins as Ion Channels: Current and Potential Research, Front. Immunol, doi:10.3389/fimmu.2020.573339
Mcgrath, Xue, Dillen, Oldfield, Assad-Garcia et al., SARS-CoV-2 variant spike and accessory gene mutations alter pathogenesis, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2204717119
Meinhardt, Radke, Dittmayer, Franz, Thomas et al., Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19, Nat. Neurosci, doi:10.1038/s41593-020-00758-5
Miller, Houlihan, Matamala, Cabezas-Bratesco, Lee et al., The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins, eLife, doi:10.7554/eLife.84477
Minakshi, Padhan, The YXXPhi motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport, Virol. J, doi:10.1186/1743-422X-11-75
Miura, Suzuki, Ishida, Arakawa, Wu et al., Distinct motifs in the E protein are required for SARS-CoV-2 virus particle formation and lysosomal deacidification in host cells, J. Virol, doi:10.1128/jvi.00426-23
Nagy, Pongor, Gyorffy, Different mutations in SARS-CoV-2 associate with severe and mild outcome, Int. J. Antimicrob. Agents, doi:10.1016/j.ijantimicag.2020.106272
Nieto-Torres, Dediego, Verdia-Baguena, Jimenez-Guardeno, Regla-Nava et al., Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis, PLoS Pathog, doi:10.1371/journal.ppat.1004077
Nieto-Torres, Verdia-Baguena, Jimenez-Guardeno, Regla-Nava, Castano-Rodriguez et al., Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome, Virology, doi:10.1016/j.virol.2015.08.010
Nieva, Madan, Carrasco, Viroporins: Structure and biological functions, Nat. Rev. Microbiol, doi:10.1038/nrmicro2820
Nishimura, Balch, A di-acidic signal required for selective export from the endoplasmic reticulum, Science, doi:10.1126/science.277.5325.556
Nkeze, Li, Benko, Li, Zhao, Molecular characterization of HIV-1 genome in fission yeast Schizosaccharomyces pombe, Cell Biosci, doi:10.1186/s13578-015-0037-7
Oostra, De Haan, De Groot, Rottier, Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M, J. Virol, doi:10.1128/JVI.80.5.2326-2336.2006
Padhan, Tanwar, Hussain, Hui, Lee et al., Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin, J. Gen. Virol, doi:10.1099/vir.0.82856-0
Papadimitriou, Drachenberg, Kleiner, Choudhri, Haririan et al., Tubular Epithelial and Peritubular Capillary Endothelial Injury in COVID-19 AKI, Kidney Int. Rep, doi:10.1016/j.ekir.2020.10.029
Paudel, Shaikh, Chakraborti, Kumari, Aledo-Serrano et al., HMGB1: A Common Biomarker and Potential Target for TBI, Neuroinflammation, Epilepsy, and Cognitive Dysfunction, Front. Neurosci, doi:10.3389/fnins.2018.00628
Peng, Du, Son, Diamond, HIF-1alpha is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2106017118
Puelles, Lutgehetmann, Lindenmeyer, Sperhake, Wong et al., Multiorgan and Renal Tropism of SARS-CoV-2, N. Engl. J. Med, doi:10.1056/NEJMc2011400
Qin, Zhao, Liu, Zhang, Yang et al., RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models, Cell Discov, doi:10.1038/s41421-022-00450-x
Ratajczak, Kucia, SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine "storm" and risk factor for damage of hematopoietic stem cells, Leukemia, doi:10.1038/s41375-020-0887-9
Ren, Shu, Wu, Mu, Wang et al., The ORF3a protein of SARS-CoV-2 induces apoptosis in cells, Cell Mol. Immunol, doi:10.1038/s41423-020-0485-9
Rice, Kimata, SARS-CoV-2 likely targets cellular PDZ proteins: A common tactic of pathogenic viruses, Future Virol, doi:10.2217/fvl-2020-0365
Rubin, From Positive to Negative to Positive Again-The Mystery of Why COVID-19 Rebounds in Some Patients Who Take Paxlovid, JAMA, doi:10.1001/jama.2022.9925
Samandari, Ongalo, Mccarthy, Biegon, Madiega et al., Prevalence and functional profile of SARS-CoV-2 T cells in asymptomatic Kenyan adults, J. Clin. Investig, doi:10.1172/JCI170011
Sayah, Berkane, Guermache, Sabri, Lakhal et al., Interleukin-6, procalcitonin and neutrophil-to-lymphocyte ratio: Potential immune-inflammatory parameters to identify severe and fatal forms of COVID-19, Cytokine, doi:10.1016/j.cyto.2021.155428
Schiff, Hadker, Weiser, Rausch, A literature review of the feasibility of glial fibrillary acidic protein as a biomarker for stroke and traumatic brain injury, Mol. Diagn. Ther, doi:10.1007/BF03256432
Service, Bad news for Paxlovid? Resistance may be coming, Science, doi:10.1126/science.add8037
Sharfuddin, Molitoris, Pathophysiology of ischemic acute kidney injury, Nat. Rev. Nephrol, doi:10.1038/nrneph.2011.16
Shariq, Malik, Sheikh, Hasnain, Ehtesham, Regulation of autophagy by SARS-CoV-2: The multifunctional contributions of ORF3a, J. Med. Virol, doi:10.1002/jmv.28959
Shi, Zhao, Wang, Shi, Wang et al., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death, Nature, doi:10.1038/nature15514
Silvas, Vasquez, Park, Chiem, Allue-Guardia et al., Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice, J. Virol, doi:10.1128/JVI.00402-21
Siu, Yuen, Castano-Rodriguez, Ye, Yeung et al., Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC, FASEB J, doi:10.1096/fj.201802418R
Spudich, Nath, Nervous system consequences of COVID-19, Science, doi:10.1126/science.abm2052
Stewart, Palmulli, Johansen, Mcgovern, Shehata et al., Tetherin antagonism by SARS-CoV-2 ORF3a and spike protein enhances virus release, EMBO Rep, doi:10.15252/embr.202357224
Su, Wang, Yoo, Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2, Sci. Rep, doi:10.1038/s41598-021-92941-2
Su, Yang, Wan, Yi, Tang et al., Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China, Kidney Int, doi:10.1016/j.kint.2020.04.003
Tan, Linster, Tan, Le Bert, Chia et al., Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients, Cell Rep, doi:10.1016/j.celrep.2021.108728
Tan, Teng, Shen, Tan, Goh et al., A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis, J. Virol, doi:10.1128/JVI.78.13.6723-6734.2004
Tan, Zhao, Li, Ning, Huang et al., HMGB1 mediates cognitive impairment caused by the NLRP3 inflammasome in the late stage of traumatic brain injury, J. Neuroinflamm, doi:10.1186/s12974-021-02274-0
Treeza, Augustine, Mathew, Kanthlal, Panonummal, Targeting Viral ORF3a Protein: A New Approach to Mitigate COVID-19 Induced Immune Cell Apoptosis and Associated Respiratory Complications, Adv. Pharm. Bull, doi:10.34172/apb.2023.069
Tzou, Tao, Nouhin, Rhee, Hu et al., Coronavirus Antiviral Research Database (CoV-RDB): An Online Database Designed to Facilitate Comparisons between Candidate Anti-Coronavirus Compounds, Viruses, doi:10.3390/v12091006
Tzou, Tao, Pond, Shafer, Coronavirus Resistance Database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons, PLoS ONE, doi:10.1371/journal.pone.0261045
Valle, Kim-Schulze, Huang, Beckmann, Nirenberg et al., An inflammatory cytokine signature predicts COVID-19 severity and survival, Nat. Med, doi:10.1038/s41591-020-1051-9
Van Den Berg, Te Velde, Severe COVID-19: NLRP3 Inflammasome Dysregulated, Front. Immunol, doi:10.3389/fimmu.2020.01580
Wang, Zhang, Du, Du, Zhao et al., Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial, Lancet, doi:10.1016/S0140-6736(20)31022-9
Weinreich, Sivapalasingam, Norton, Ali, Gao et al., REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19, N. Engl. J. Med, doi:10.1056/NEJMoa2035002
Xie, Edwards, Adam, Leung, Tsang et al., Resurgence of Omicron BA.2 in SARS-CoV-2 infection-naive Hong Kong, Nat. Commun
Xu, Akinyemi, Chitre, Loeb, Lednicky et al., SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway, Virology, doi:10.1016/j.virol.2022.01.003
Xu, Xie, Al-Aly, Long-term neurologic outcomes of COVID-19, Nat. Med, doi:10.1038/s41591-022-02001-z
Yalcinkaya, Liu, Islam, Kotini, Gusarova et al., Modulation of the NLRP3 inflammasome by SARS-CoV-2 Envelope protein, Sci. Rep, doi:10.1038/s41598-021-04133-7
Yang, Nkeze, Zhao, Effects of HIV-1 protease on cellular functions and their potential applications in antiretroviral therapy, Cell Biosci, doi:10.1186/2045-3701-2-32
Yang, Wang, Andersson, Targeting Inflammation Driven by HMGB1, Front. Immunol, doi:10.3389/fimmu.2020.00484
Yang, Wang, Kouadir, Song, Shi, Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors, Cell Death Dis, doi:10.1038/s41419-019-1413-8
Ye, Park, Chiem, Dravid, Allue-Guardia et al., Immunization with Recombinant Accessory Protein-Deficient SARS-CoV-2 Protects against Lethal Challenge and Viral Transmission, Microbiol. Spectr, doi:10.1128/spectrum.00653-23
Yount, Roberts, Sims, Deming, Frieman et al., Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice, J. Virol, doi:10.1128/JVI.79.23.14909-14922.2005
Zhang, Cruz-Cosme, Zhang, Liu, Tang et al., Endoplasmic reticulum-associated SARS-CoV-2 ORF3a elicits heightened cytopathic effects despite robust ER-associated degradation, mBio, doi:10.1128/mbio.03030-23
Zhang, Ejikemeuwa, Gerzanich, Nasr, Tang et al., Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19, Front. Microbiol, doi:10.3389/fmicb.2022.854567
Zhang, Li, Cruz Cosme, Gerzanich, Tang et al., Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets, mBio, doi:10.1128/mbio.00169-22
Zhang, Li, Kawashima, Nasr, Xue et al., Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model, Pathogens, doi:10.3390/pathogens11070804
Zhang, Liu, Liu, Bailey, Plante et al., A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence, Cell, doi:10.1016/j.cell.2021.02.044
Zhang, Nasr, Zhao, None
Zhang, Sun, Pei, Mao, Zhao et al., The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes, Cell Discov, doi:10.1038/s41421-021-00268-z
Zhang, Vernon, Li, Benko, Amoroso et al., Single-Agent and Fixed-Dose Combination HIV-1 Protease Inhibitor Drugs in Fission Yeast (Schizosaccharomyces pombe), Pathogens, doi:10.3390/pathogens10070804
Zhang, Wang, Ping, Yu, Qian et al., The ns12.9 Accessory Protein of Human Coronavirus OC43 Is a Viroporin Involved in Virion Morphogenesis and Pathogenesis, J. Virol, doi:10.1128/JVI.01986-15
Zhang, Wu, Ma, Wei, Garstka et al., The C5a/C5aR2 axis promotes renal inflammation and tissue damage, JCI Insight, doi:10.1172/jci.insight.134081
Zhang, Wu, Yao, Zhang, Zhou et al., Pyroptotic macrophages stimulate the SARS-CoV-2-associated cytokine storm, Cell Mol. Immunol, doi:10.1038/s41423-021-00665-0
Zhao, Elder, Chen, Cao, Fission yeast expression vectors adapted for positive identification of gene insertion and green fluorescent protein fusion, Biotechniques, doi:10.2144/98253st06
Zhao, Elder, Yeast perspectives on HIV-1 Vpr, Front. Biosci, doi:10.2741/zhao
Zhao, Yeast for virus research, Microb. Cell, doi:10.15698/mic2017.10.592
Zhong, Guo, Yang, Peng, Xie et al., Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients, J. Gen. Virol, doi:10.1099/vir.0.81078-0
Zhu, Byrnes, Lee, Tuymetova, Duffy et al., SARS-CoV-2 ORF3a expression in brain disrupts the autophagy-lysosomal pathway, impairs sphingolipid homeostasis, and drives neuropathogenesis, FASEB J, doi:10.1096/fj.202300149R
Zhu, Liu, Li, Lei, Wu et al., Age-and Severity-Associated Humoral Immunity Response in COVID-19 Patients: A Cohort Study from Wuhan, China, J. Clin. Med, doi:10.3390/jcm11195974
DOI record:
{
"DOI": "10.3390/pathogens13010075",
"ISSN": [
"2076-0817"
],
"URL": "http://dx.doi.org/10.3390/pathogens13010075",
"abstract": "<jats:p>The COVID-19 pandemic caused by SARS-CoV-2 has posed unparalleled challenges due to its rapid transmission, ability to mutate, high mortality and morbidity, and enduring health complications. Vaccines have exhibited effectiveness, but their efficacy diminishes over time while new variants continue to emerge. Antiviral medications offer a viable alternative, but their success has been inconsistent. Therefore, there remains an ongoing need to identify innovative antiviral drugs for treating COVID-19 and its post-infection complications. The ORF3a (open reading frame 3a) protein found in SARS-CoV-2, represents a promising target for antiviral treatment due to its multifaceted role in viral pathogenesis, cytokine storms, disease severity, and mortality. ORF3a contributes significantly to viral pathogenesis by facilitating viral assembly and release, essential processes in the viral life cycle, while also suppressing the body’s antiviral responses, thus aiding viral replication. ORF3a also has been implicated in triggering excessive inflammation, characterized by NF-κB-mediated cytokine production, ultimately leading to apoptotic cell death and tissue damage in the lungs, kidneys, and the central nervous system. Additionally, ORF3a triggers the activation of the NLRP3 inflammasome, inciting a cytokine storm, which is a major contributor to the severity of the disease and subsequent mortality. As with the spike protein, ORF3a also undergoes mutations, and certain mutant variants correlate with heightened disease severity in COVID-19. These mutations may influence viral replication and host cellular inflammatory responses. While establishing a direct link between ORF3a and mortality is difficult, its involvement in promoting inflammation and exacerbating disease severity likely contributes to higher mortality rates in severe COVID-19 cases. This review offers a comprehensive and detailed exploration of ORF3a’s potential as an innovative antiviral drug target. Additionally, we outline potential strategies for discovering and developing ORF3a inhibitor drugs to counteract its harmful effects, alleviate tissue damage, and reduce the severity of COVID-19 and its lingering complications.</jats:p>",
"alternative-id": [
"pathogens13010075"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-2875-9743",
"affiliation": [
{
"name": "Department of Pathology, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
}
],
"authenticated-orcid": false,
"family": "Zhang",
"given": "Jiantao",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD 21201, USA"
}
],
"family": "Hom",
"given": "Kellie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pathology, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
}
],
"family": "Zhang",
"given": "Chenyu",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3322-3716",
"affiliation": [
{
"name": "Drug Development and Clinical Sciences Branch, Division of AIDS, NIAID, National Institutes of Health, Bethesda, MD 20892, USA"
}
],
"authenticated-orcid": false,
"family": "Nasr",
"given": "Mohamed",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6714-1946",
"affiliation": [
{
"name": "Department of Neurosurgery, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
}
],
"authenticated-orcid": false,
"family": "Gerzanich",
"given": "Volodymyr",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5847-3260",
"affiliation": [
{
"name": "Department of Veterinary Medicine, University of Maryland, College Park, MD 20742, USA"
}
],
"authenticated-orcid": false,
"family": "Zhang",
"given": "Yanjin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6487-2356",
"affiliation": [
{
"name": "Department of Microbiology, Howard University College of Medicine, Washington, DC 20059, USA"
}
],
"authenticated-orcid": false,
"family": "Tang",
"given": "Qiyi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD 21201, USA"
}
],
"family": "Xue",
"given": "Fengtian",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5373-1988",
"affiliation": [
{
"name": "Department of Neurosurgery, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
},
{
"name": "Research & Development Service, VA Maryland Health Care System, Baltimore, MD 21201, USA"
}
],
"authenticated-orcid": false,
"family": "Simard",
"given": "J. Marc",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3424-2852",
"affiliation": [
{
"name": "Department of Pathology, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
},
{
"name": "Research & Development Service, VA Maryland Health Care System, Baltimore, MD 21201, USA"
},
{
"name": "Department of Microbiology-Immunology, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
},
{
"name": "Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
},
{
"name": "Institute of Global Health, University of Maryland School of Medicine, Baltimore, MD 21201, USA"
}
],
"authenticated-orcid": false,
"family": "Zhao",
"given": "Richard Y.",
"sequence": "additional"
}
],
"container-title": "Pathogens",
"container-title-short": "Pathogens",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
1,
15
]
],
"date-time": "2024-01-15T13:52:03Z",
"timestamp": 1705326723000
},
"deposited": {
"date-parts": [
[
2024,
1,
15
]
],
"date-time": "2024-01-15T14:41:56Z",
"timestamp": 1705329716000
},
"indexed": {
"date-parts": [
[
2024,
1,
16
]
],
"date-time": "2024-01-16T00:14:19Z",
"timestamp": 1705364059682
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
1,
14
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
1,
14
]
],
"date-time": "2024-01-14T00:00:00Z",
"timestamp": 1705190400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-0817/13/1/75/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "75",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2024,
1,
14
]
]
},
"published-online": {
"date-parts": [
[
2024,
1,
14
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.3390/vaccines10050755",
"doi-asserted-by": "crossref",
"key": "ref_1",
"unstructured": "Alcendor, D.J., Matthews-Juarez, P., Smoot, D., Hildreth, J.E.K., Lamar, K., Tabatabai, M., Wilus, D., and Juarez, P.D. (2022). Breakthrough COVID-19 Infections in the US: Implications for Prolonging the Pandemic. Vaccines, 10."
},
{
"DOI": "10.1001/jama.2022.9925",
"article-title": "From Positive to Negative to Positive Again-The Mystery of Why COVID-19 Rebounds in Some Patients Who Take Paxlovid",
"author": "Rubin",
"doi-asserted-by": "crossref",
"first-page": "2380",
"journal-title": "JAMA",
"key": "ref_2",
"volume": "327",
"year": "2022"
},
{
"DOI": "10.1126/science.add8037",
"article-title": "Bad news for Paxlovid? Resistance may be coming",
"author": "Service",
"doi-asserted-by": "crossref",
"first-page": "138",
"journal-title": "Science",
"key": "ref_3",
"volume": "377",
"year": "2022"
},
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the Treatment of COVID-19—Final Report",
"author": "Beigel",
"doi-asserted-by": "crossref",
"first-page": "1813",
"journal-title": "N. Engl. J. Med.",
"key": "ref_4",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"journal-title": "Cell",
"key": "ref_5",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors",
"author": "Jin",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "ref_6",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)31022-9",
"article-title": "Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1569",
"journal-title": "Lancet",
"key": "ref_7",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2021436",
"article-title": "Dexamethasone in Hospitalized Patients with COVID-19",
"author": "Group",
"doi-asserted-by": "crossref",
"first-page": "693",
"journal-title": "N. Engl. J. Med.",
"key": "ref_8",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.1056/NEJMoa2035002",
"article-title": "REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19",
"author": "Weinreich",
"doi-asserted-by": "crossref",
"first-page": "238",
"journal-title": "N. Engl. J. Med.",
"key": "ref_9",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.15698/mic2017.10.592",
"article-title": "Yeast for virus research",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "311",
"journal-title": "Microb. Cell",
"key": "ref_10",
"volume": "4",
"year": "2017"
},
{
"DOI": "10.1007/978-1-4939-7546-4_5",
"article-title": "Molecular Cloning and Characterization of Small Viral Genome in Fission Yeast",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "47",
"journal-title": "Methods Mol. Biol.",
"key": "ref_11",
"volume": "1721",
"year": "2018"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "crossref",
"first-page": "459",
"journal-title": "Nature",
"key": "ref_12",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.3201/eid2606.200516",
"article-title": "Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States",
"author": "Harcourt",
"doi-asserted-by": "crossref",
"first-page": "1266",
"journal-title": "Emerg. Infect. Dis.",
"key": "ref_13",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.2144/98253st06",
"article-title": "Fission yeast expression vectors adapted for positive identification of gene insertion and green fluorescent protein fusion",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "438",
"journal-title": "Biotechniques",
"key": "ref_14",
"volume": "25",
"year": "1998"
},
{
"DOI": "10.1186/s13578-015-0037-7",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Nkeze, J., Li, L., Benko, Z., Li, G., and Zhao, R.Y. (2015). Molecular characterization of HIV-1 genome in fission yeast Schizosaccharomyces pombe. Cell Biosci., 5."
},
{
"article-title": "Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast",
"author": "Li",
"first-page": "E376",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_16",
"volume": "114",
"year": "2017"
},
{
"DOI": "10.1101/2021.11.23.469747",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Zhang, J., Li, Q., Cruz Cosme, R.S., Gerzanich, V., Tang, Q., Simard, J.M., and Zhao, R.Y. (2021). Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets. mBio, 13."
},
{
"DOI": "10.3389/fmicb.2022.854567",
"doi-asserted-by": "crossref",
"key": "ref_18",
"unstructured": "Zhang, J., Ejikemeuwa, A., Gerzanich, V., Nasr, M., Tang, Q., Simard, J.M., and Zhao, R.Y. (2022). Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19. Front. Microbiol., 13."
},
{
"DOI": "10.1038/s41590-020-0773-7",
"article-title": "ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection",
"author": "Hachim",
"doi-asserted-by": "crossref",
"first-page": "1293",
"journal-title": "Nat. Immunol.",
"key": "ref_19",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1128/Spectrum.01416-21",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Camerini, D., Randall, A.Z., Trappl-Kimmons, K., Oberai, A., Hung, C., Edgar, J., Shandling, A., Huynh, V., Teng, A.A., and Hermanson, G. (2021). Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray. Microbiol. Spectr., 9."
},
{
"DOI": "10.1086/422753",
"article-title": "Disease-specific B Cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "797",
"journal-title": "J. Infect. Dis.",
"key": "ref_21",
"volume": "190",
"year": "2004"
},
{
"DOI": "10.1099/vir.0.81078-0",
"article-title": "Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients",
"author": "Zhong",
"doi-asserted-by": "crossref",
"first-page": "369",
"journal-title": "J. Gen. Virol.",
"key": "ref_22",
"volume": "87",
"year": "2006"
},
{
"DOI": "10.3390/jcm11195974",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Zhu, A., Liu, M., Li, Y., Lei, Q., Wu, Q., Lin, M., Lai, D., Lu, L., Yu, S., and Guo, S. (2022). Age- and Severity-Associated Humoral Immunity Response in COVID-19 Patients: A Cohort Study from Wuhan, China. J. Clin. Med., 11."
},
{
"DOI": "10.1016/j.cell.2020.05.015",
"article-title": "Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals",
"author": "Grifoni",
"doi-asserted-by": "crossref",
"first-page": "1489",
"journal-title": "Cell",
"key": "ref_24",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.celrep.2021.108728",
"article-title": "Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients",
"author": "Tan",
"doi-asserted-by": "crossref",
"first-page": "108728",
"journal-title": "Cell Rep.",
"key": "ref_25",
"volume": "34",
"year": "2021"
},
{
"DOI": "10.1172/JCI170011",
"article-title": "Prevalence and functional profile of SARS-CoV-2 T cells in asymptomatic Kenyan adults",
"author": "Samandari",
"doi-asserted-by": "crossref",
"first-page": "13",
"journal-title": "J. Clin. Investig.",
"key": "ref_26",
"volume": "133",
"year": "2023"
},
{
"DOI": "10.1128/JVI.01986-15",
"article-title": "The ns12.9 Accessory Protein of Human Coronavirus OC43 Is a Viroporin Involved in Virion Morphogenesis and Pathogenesis",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "11383",
"journal-title": "J. Virol.",
"key": "ref_27",
"volume": "89",
"year": "2015"
},
{
"DOI": "10.1038/s41594-021-00619-0",
"article-title": "Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs",
"author": "Kern",
"doi-asserted-by": "crossref",
"first-page": "573",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_28",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2020.573339",
"article-title": "Coronavirus Proteins as Ion Channels: Current and Potential Research",
"author": "McClenaghan",
"doi-asserted-by": "crossref",
"first-page": "573339",
"journal-title": "Front. Immunol.",
"key": "ref_29",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1128/mSystems.00266-20",
"article-title": "SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis",
"author": "Issa",
"doi-asserted-by": "crossref",
"first-page": "e00266-20",
"journal-title": "mSystems",
"key": "ref_30",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.3389/fcell.2022.1011221",
"doi-asserted-by": "crossref",
"key": "ref_31",
"unstructured": "Cruz-Cosme, R., Zhang, J., Liu, D., Mahase, V., Sallapalli, B.T., Chang, P., Zhang, Y., Teng, S., Zhao, R.Y., and Tang, Q. (2022). A novel diG motif in ORF3a protein of SARS-CoV-2 for intracellular transport. Front. Cell Dev. Biol., 10."
},
{
"DOI": "10.1099/vir.0.82856-0",
"article-title": "Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin",
"author": "Padhan",
"doi-asserted-by": "crossref",
"first-page": "3067",
"journal-title": "J. Gen. Virol.",
"key": "ref_32",
"volume": "88",
"year": "2007"
},
{
"DOI": "10.1128/JVI.78.13.6723-6734.2004",
"article-title": "A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis",
"author": "Tan",
"doi-asserted-by": "crossref",
"first-page": "6723",
"journal-title": "J. Virol.",
"key": "ref_33",
"volume": "78",
"year": "2004"
},
{
"DOI": "10.1186/1743-422X-11-75",
"article-title": "The YXXPhi motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport",
"author": "Minakshi",
"doi-asserted-by": "crossref",
"first-page": "75",
"journal-title": "Virol. J.",
"key": "ref_34",
"volume": "11",
"year": "2014"
},
{
"DOI": "10.1016/j.ijbiomac.2020.12.142",
"article-title": "SARS-CoV-2 ORF3a: Mutability and function",
"author": "Bianchi",
"doi-asserted-by": "crossref",
"first-page": "820",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_35",
"volume": "170",
"year": "2021"
},
{
"DOI": "10.3390/biom12081052",
"doi-asserted-by": "crossref",
"key": "ref_36",
"unstructured": "Kakkanas, A., Karamichali, E., Koufogeorgou, E.I., Kotsakis, S.D., Georgopoulou, U., and Foka, P. (2022). Targeting the YXXPhi Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions. Biomolecules, 12."
},
{
"DOI": "10.1096/fj.201802418R",
"article-title": "Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC",
"author": "Siu",
"doi-asserted-by": "crossref",
"first-page": "8865",
"journal-title": "FASEB J.",
"key": "ref_37",
"volume": "33",
"year": "2019"
},
{
"article-title": "Mechanism of inflammasome activation by SARS coronavirus 3a protein: Abridged secondary publication",
"author": "Jin",
"first-page": "33",
"journal-title": "Hong Kong Med. J.",
"key": "ref_38",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2023.1064293",
"article-title": "SARS-CoV-2 viroporins activate the NLRP3-inflammasome by the mitochondrial permeability transition pore",
"author": "Guarnieri",
"doi-asserted-by": "crossref",
"first-page": "1064293",
"journal-title": "Front. Immunol.",
"key": "ref_39",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.ymthe.2022.12.008",
"article-title": "SARS-CoV-2 viral protein ORF3A injures renal tubules by interacting with TRIM59 to induce STAT3 activation",
"author": "Cai",
"doi-asserted-by": "crossref",
"first-page": "774",
"journal-title": "Mol. Ther.",
"key": "ref_40",
"volume": "31",
"year": "2023"
},
{
"DOI": "10.1096/fj.202300149R",
"article-title": "SARS-CoV-2 ORF3a expression in brain disrupts the autophagy-lysosomal pathway, impairs sphingolipid homeostasis, and drives neuropathogenesis",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "e22919",
"journal-title": "FASEB J.",
"key": "ref_41",
"volume": "37",
"year": "2023"
},
{
"DOI": "10.1046/j.1523-1755.2002.00433.x",
"article-title": "Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury",
"author": "Han",
"doi-asserted-by": "crossref",
"first-page": "237",
"journal-title": "Kidney Int.",
"key": "ref_42",
"volume": "62",
"year": "2002"
},
{
"DOI": "10.1038/nature21029",
"article-title": "Neurotoxic reactive astrocytes are induced by activated microglia",
"author": "Liddelow",
"doi-asserted-by": "crossref",
"first-page": "481",
"journal-title": "Nature",
"key": "ref_43",
"volume": "541",
"year": "2017"
},
{
"DOI": "10.1007/BF03256432",
"article-title": "A literature review of the feasibility of glial fibrillary acidic protein as a biomarker for stroke and traumatic brain injury",
"author": "Schiff",
"doi-asserted-by": "crossref",
"first-page": "79",
"journal-title": "Mol. Diagn. Ther.",
"key": "ref_44",
"volume": "16",
"year": "2012"
},
{
"DOI": "10.1186/s13578-021-00643-z",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Brant, A.C., Tian, W., Majerciak, V., Yang, W., and Zheng, Z.M. (2021). SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci., 11."
},
{
"DOI": "10.1016/j.celrep.2023.112835",
"article-title": "Super-resolution proximity labeling reveals anti-viral protein network and its structural changes against SARS-CoV-2 viral proteins",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "112835",
"journal-title": "Cell Rep.",
"key": "ref_46",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1128/mbio.03030-23",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Zhang, J., Cruz-Cosme, R., Zhang, C., Liu, D., Tang, Q., and Zhao, R.Y. (2023). Endoplasmic reticulum-associated SARS-CoV-2 ORF3a elicits heightened cytopathic effects despite robust ER-associated degradation. mBio."
},
{
"DOI": "10.1126/science.277.5325.556",
"article-title": "A di-acidic signal required for selective export from the endoplasmic reticulum",
"author": "Nishimura",
"doi-asserted-by": "crossref",
"first-page": "556",
"journal-title": "Science",
"key": "ref_48",
"volume": "277",
"year": "1997"
},
{
"DOI": "10.1128/JVI.80.5.2326-2336.2006",
"article-title": "Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M",
"author": "Oostra",
"doi-asserted-by": "crossref",
"first-page": "2326",
"journal-title": "J. Virol.",
"key": "ref_49",
"volume": "80",
"year": "2006"
},
{
"DOI": "10.1016/j.biocel.2022.106185",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Breitinger, U., Farag, N.S., Sticht, H., and Breitinger, H.G. (2022). Viroporins: Structure, function, and their role in the life cycle of SARS-CoV-2. Int. J. Biochem. Cell Biol., 145."
},
{
"DOI": "10.1016/j.virol.2022.01.003",
"article-title": "SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "13",
"journal-title": "Virology",
"key": "ref_51",
"volume": "568",
"year": "2022"
},
{
"DOI": "10.1038/s41598-023-31764-9",
"article-title": "Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites",
"author": "Fam",
"doi-asserted-by": "crossref",
"first-page": "5328",
"journal-title": "Sci. Rep.",
"key": "ref_52",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1038/nrmicro2820",
"article-title": "Viroporins: Structure and biological functions",
"author": "Nieva",
"doi-asserted-by": "crossref",
"first-page": "563",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_53",
"volume": "10",
"year": "2012"
},
{
"DOI": "10.7554/eLife.84477",
"article-title": "The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins",
"author": "Miller",
"doi-asserted-by": "crossref",
"first-page": "e84477",
"journal-title": "eLife",
"key": "ref_54",
"volume": "12",
"year": "2023"
},
{
"DOI": "10.3390/v15112229",
"doi-asserted-by": "crossref",
"key": "ref_55",
"unstructured": "Busscher, B.M., Befekadu, H.B., Liu, Z., and Xiao, T.S. (2023). SARS-CoV-2 ORF3a-Mediated NF-kappaB Activation Is Not Dependent on TRAF-Binding Sequence. Viruses, 15."
},
{
"DOI": "10.1111/febs.15881",
"article-title": "Host PDZ-containing proteins targeted by SARS-CoV-2",
"author": "Durbesson",
"doi-asserted-by": "crossref",
"first-page": "5148",
"journal-title": "FEBS J.",
"key": "ref_56",
"volume": "288",
"year": "2021"
},
{
"article-title": "Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis",
"author": "Honrubia",
"first-page": "10",
"journal-title": "mBio",
"key": "ref_57",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.2217/fvl-2020-0365",
"article-title": "SARS-CoV-2 likely targets cellular PDZ proteins: A common tactic of pathogenic viruses",
"author": "Rice",
"doi-asserted-by": "crossref",
"first-page": "375",
"journal-title": "Future Virol.",
"key": "ref_58",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1128/jvi.00426-23",
"article-title": "Distinct motifs in the E protein are required for SARS-CoV-2 virus particle formation and lysosomal deacidification in host cells",
"author": "Miura",
"doi-asserted-by": "crossref",
"first-page": "e0042623",
"journal-title": "J. Virol.",
"key": "ref_59",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1128/mbio.03136-22",
"doi-asserted-by": "crossref",
"key": "ref_60",
"unstructured": "Honrubia, J.M., Gutierrez-Alvarez, J., Sanz-Bravo, A., Gonzalez-Miranda, E., Munoz-Santos, D., Castano-Rodriguez, C., Wang, L., Villarejo-Torres, M., Ripoll-Gomez, J., and Esteban, A. (2023). SARS-CoV-2-Mediated Lung Edema and Replication Are Diminished by Cystic Fibrosis Transmembrane Conductance Regulator Modulators. mBio, 14."
},
{
"DOI": "10.1016/j.cell.2021.02.044",
"article-title": "A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "2229",
"journal-title": "Cell",
"key": "ref_61",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.devcel.2021.10.006",
"article-title": "ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "3250",
"journal-title": "Dev. Cell",
"key": "ref_62",
"volume": "56",
"year": "2021"
},
{
"DOI": "10.1073/pnas.0605402103",
"article-title": "Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "12540",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_63",
"volume": "103",
"year": "2006"
},
{
"DOI": "10.1016/j.cell.2020.10.039",
"article-title": "beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway",
"author": "Ghosh",
"doi-asserted-by": "crossref",
"first-page": "1520",
"journal-title": "Cell",
"key": "ref_64",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.15252/embr.202357224",
"article-title": "Tetherin antagonism by SARS-CoV-2 ORF3a and spike protein enhances virus release",
"author": "Stewart",
"doi-asserted-by": "crossref",
"first-page": "e57224",
"journal-title": "EMBO Rep.",
"key": "ref_65",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/j.molcel.2021.04.008",
"article-title": "Functional landscape of SARS-CoV-2 cellular restriction",
"author": "Lewinski",
"doi-asserted-by": "crossref",
"first-page": "2656",
"journal-title": "Mol. Cell",
"key": "ref_66",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1128/JVI.79.23.14909-14922.2005",
"article-title": "Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice",
"author": "Yount",
"doi-asserted-by": "crossref",
"first-page": "14909",
"journal-title": "J. Virol.",
"key": "ref_67",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1016/j.antiviral.2006.10.008",
"article-title": "Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S",
"author": "Akerstrom",
"doi-asserted-by": "crossref",
"first-page": "219",
"journal-title": "Antivir. Res.",
"key": "ref_68",
"volume": "73",
"year": "2007"
},
{
"DOI": "10.1128/JVI.00402-21",
"article-title": "Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice",
"author": "Silvas",
"doi-asserted-by": "crossref",
"first-page": "e0040221",
"journal-title": "J. Virol.",
"key": "ref_69",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1038/s41467-022-31930-z",
"article-title": "A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "4337",
"journal-title": "Nat. Commun.",
"key": "ref_70",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1093/infdis/jiad320",
"article-title": "Differential Severe Acute Respiratory Syndrome Coronavirus 2-Specific Humoral Response in Inactivated Virus-Vaccinated, Convalescent, and Breakthrough-Infected Subjects",
"author": "Duarte",
"doi-asserted-by": "crossref",
"first-page": "857",
"journal-title": "J. Infect. Dis.",
"key": "ref_71",
"volume": "228",
"year": "2023"
},
{
"DOI": "10.1073/pnas.2204717119",
"article-title": "SARS-CoV-2 variant spike and accessory gene mutations alter pathogenesis",
"author": "McGrath",
"doi-asserted-by": "crossref",
"first-page": "e2204717119",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_72",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1128/spectrum.00653-23",
"doi-asserted-by": "crossref",
"key": "ref_73",
"unstructured": "Ye, C., Park, J.G., Chiem, K., Dravid, P., Allue-Guardia, A., Garcia-Vilanova, A., Pino Tamayo, P., Shivanna, V., Kapoor, A., and Walter, M.R. (2023). Immunization with Recombinant Accessory Protein-Deficient SARS-CoV-2 Protects against Lethal Challenge and Viral Transmission. Microbiol. Spectr., 11."
},
{
"DOI": "10.1038/s41423-021-00665-0",
"article-title": "Pyroptotic macrophages stimulate the SARS-CoV-2-associated cytokine storm",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "1305",
"journal-title": "Cell Mol. Immunol.",
"key": "ref_74",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1038/s41375-020-0887-9",
"article-title": "SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells",
"author": "Ratajczak",
"doi-asserted-by": "crossref",
"first-page": "1726",
"journal-title": "Leukemia",
"key": "ref_75",
"volume": "34",
"year": "2020"
},
{
"DOI": "10.1002/jmv.26232",
"article-title": "The cytokine storm and COVID-19",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "250",
"journal-title": "J. Med. Virol.",
"key": "ref_76",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2020.01518",
"article-title": "Targeting the NLRP3 Inflammasome in Severe COVID-19",
"author": "Freeman",
"doi-asserted-by": "crossref",
"first-page": "1518",
"journal-title": "Front. Immunol.",
"key": "ref_77",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2020.01580",
"article-title": "Severe COVID-19: NLRP3 Inflammasome Dysregulated",
"doi-asserted-by": "crossref",
"first-page": "1580",
"journal-title": "Front. Immunol.",
"key": "ref_78",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.jaci.2022.05.029",
"article-title": "Spontaneous NLRP3 inflammasome-driven IL1-beta secretion is induced in severe COVID-19 patients and responds to anakinra treatment",
"author": "Bertoni",
"doi-asserted-by": "crossref",
"first-page": "796",
"journal-title": "J. Allergy Clin. Immunol.",
"key": "ref_79",
"volume": "150",
"year": "2022"
},
{
"DOI": "10.1111/imr.12287",
"article-title": "Pyroptotic cell death defends against intracellular pathogens",
"author": "Jorgensen",
"doi-asserted-by": "crossref",
"first-page": "130",
"journal-title": "Immunol. Rev.",
"key": "ref_80",
"volume": "265",
"year": "2015"
},
{
"DOI": "10.1038/s41419-019-1413-8",
"article-title": "Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "128",
"journal-title": "Cell Death Dis.",
"key": "ref_81",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1038/nature15541",
"article-title": "Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling",
"author": "Kayagaki",
"doi-asserted-by": "crossref",
"first-page": "666",
"journal-title": "Nature",
"key": "ref_82",
"volume": "526",
"year": "2015"
},
{
"DOI": "10.1038/nature15514",
"article-title": "Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death",
"author": "Shi",
"doi-asserted-by": "crossref",
"first-page": "660",
"journal-title": "Nature",
"key": "ref_83",
"volume": "526",
"year": "2015"
},
{
"DOI": "10.3389/fmicb.2019.00050",
"doi-asserted-by": "crossref",
"key": "ref_84",
"unstructured": "Chen, I.Y., Moriyama, M., Chang, M.F., and Ichinohe, T. (2019). Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol., 10."
},
{
"DOI": "10.1016/j.virol.2015.08.010",
"article-title": "Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome",
"author": "Torres",
"doi-asserted-by": "crossref",
"first-page": "330",
"journal-title": "Virology",
"key": "ref_85",
"volume": "485",
"year": "2015"
},
{
"DOI": "10.1371/journal.ppat.1004077",
"doi-asserted-by": "crossref",
"key": "ref_86",
"unstructured": "Nieto-Torres, J.L., DeDiego, M.L., Verdia-Baguena, C., Jimenez-Guardeno, J.M., Regla-Nava, J.A., Fernandez-Delgado, R., Castano-Rodriguez, C., Alcaraz, A., Torres, J., and Aguilella, V.M. (2014). Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog., 10."
},
{
"DOI": "10.1038/s41598-021-04133-7",
"article-title": "Modulation of the NLRP3 inflammasome by SARS-CoV-2 Envelope protein",
"author": "Yalcinkaya",
"doi-asserted-by": "crossref",
"first-page": "24432",
"journal-title": "Sci. Rep.",
"key": "ref_87",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2020.00484",
"article-title": "Targeting Inflammation Driven by HMGB1",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "484",
"journal-title": "Front. Immunol.",
"key": "ref_88",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.cyto.2021.155496",
"article-title": "Glycyrrhizin prevents SARS-CoV-2 S1 and Orf3a induced high mobility group box 1 (HMGB1) release and inhibits viral replication",
"author": "Gowda",
"doi-asserted-by": "crossref",
"first-page": "155496",
"journal-title": "Cytokine",
"key": "ref_89",
"volume": "142",
"year": "2021"
},
{
"DOI": "10.3389/fnins.2018.00628",
"article-title": "HMGB1: A Common Biomarker and Potential Target for TBI, Neuroinflammation, Epilepsy, and Cognitive Dysfunction",
"author": "Paudel",
"doi-asserted-by": "crossref",
"first-page": "628",
"journal-title": "Front. Neurosci.",
"key": "ref_90",
"volume": "12",
"year": "2018"
},
{
"DOI": "10.1073/pnas.2106017118",
"article-title": "HIF-1alpha is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes",
"author": "Peng",
"doi-asserted-by": "crossref",
"first-page": "e2106017118",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_91",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1186/s10194-021-01306-7",
"article-title": "HMGB1, NLRP3, IL-6 and ACE2 levels are elevated in COVID-19 with headache: A window to the infection-related headache mechanism",
"author": "Bolay",
"doi-asserted-by": "crossref",
"first-page": "94",
"journal-title": "J. Headache Pain",
"key": "ref_92",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1038/s41423-020-0492-x",
"article-title": "Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "992",
"journal-title": "Cell Mol. Immunol.",
"key": "ref_93",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1038/s41581-021-00452-0",
"article-title": "Pathophysiology of COVID-19-associated acute kidney injury",
"author": "Legrand",
"doi-asserted-by": "crossref",
"first-page": "751",
"journal-title": "Nat. Rev. Nephrol.",
"key": "ref_94",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1016/j.kint.2021.07.015",
"article-title": "A multi-center retrospective cohort study defines the spectrum of kidney pathology in Coronavirus 2019 Disease (COVID-19)",
"author": "May",
"doi-asserted-by": "crossref",
"first-page": "1303",
"journal-title": "Kidney Int.",
"key": "ref_95",
"volume": "100",
"year": "2021"
},
{
"DOI": "10.2215/CJN.09610620",
"article-title": "Acute Kidney Injury in a National Cohort of Hospitalized US Veterans with COVID-19",
"author": "Bowe",
"doi-asserted-by": "crossref",
"first-page": "14",
"journal-title": "Clin. J. Am. Soc. Nephrol.",
"key": "ref_96",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1681/ASN.2020050615",
"article-title": "AKI in Hospitalized Patients with COVID-19",
"author": "Chan",
"doi-asserted-by": "crossref",
"first-page": "151",
"journal-title": "J. Am. Soc. Nephrol.",
"key": "ref_97",
"volume": "32",
"year": "2021"
},
{
"DOI": "10.1186/s13613-021-00875-9",
"article-title": "Acute kidney injury in SARS-CoV2-related pneumonia ICU patients: A retrospective multicenter study",
"author": "Geri",
"doi-asserted-by": "crossref",
"first-page": "86",
"journal-title": "Ann. Intensive Care",
"key": "ref_98",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2011400",
"article-title": "Multiorgan and Renal Tropism of SARS-CoV-2",
"author": "Puelles",
"doi-asserted-by": "crossref",
"first-page": "590",
"journal-title": "N. Engl. J. Med.",
"key": "ref_99",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1007/s40620-022-01296-y",
"article-title": "Post-acute COVID-19 syndrome and kidney diseases: What do we know?",
"author": "Copur",
"doi-asserted-by": "crossref",
"first-page": "795",
"journal-title": "J. Nephrol.",
"key": "ref_100",
"volume": "35",
"year": "2022"
},
{
"DOI": "10.1021/acscentsci.0c01537",
"article-title": "CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2",
"author": "Amraei",
"doi-asserted-by": "crossref",
"first-page": "1156",
"journal-title": "ACS Cent. Sci.",
"key": "ref_101",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.3389/fcell.2022.855340",
"doi-asserted-by": "crossref",
"key": "ref_102",
"unstructured": "Kalejaiye, T.D., Bhattacharya, R., Burt, M.A., Travieso, T., Okafor, A.E., Mou, X., Blasi, M., and Musah, S. (2022). SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes. Front. Cell Dev. Biol., 10."
},
{
"DOI": "10.1681/ASN.2020081229",
"article-title": "Does SARS-CoV-2 Infect the Kidney?",
"author": "Khan",
"doi-asserted-by": "crossref",
"first-page": "2746",
"journal-title": "J. Am. Soc. Nephrol.",
"key": "ref_103",
"volume": "31",
"year": "2020"
},
{
"DOI": "10.3390/jcm10061216",
"doi-asserted-by": "crossref",
"key": "ref_104",
"unstructured": "Armaly, Z., Kinaneh, S., and Skorecki, K. (2021). Renal Manifestations of COVID-19: Physiology and Pathophysiology. J. Clin. Med., 10."
},
{
"DOI": "10.1016/j.kint.2020.04.003",
"article-title": "Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China",
"author": "Su",
"doi-asserted-by": "crossref",
"first-page": "219",
"journal-title": "Kidney Int.",
"key": "ref_105",
"volume": "98",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)31759-1",
"article-title": "SARS-CoV-2 renal tropism associates with acute kidney injury",
"author": "Braun",
"doi-asserted-by": "crossref",
"first-page": "597",
"journal-title": "Lancet",
"key": "ref_106",
"volume": "396",
"year": "2020"
},
{
"DOI": "10.1016/j.ekir.2020.10.029",
"article-title": "Tubular Epithelial and Peritubular Capillary Endothelial Injury in COVID-19 AKI",
"author": "Papadimitriou",
"doi-asserted-by": "crossref",
"first-page": "518",
"journal-title": "Kidney Int. Rep.",
"key": "ref_107",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1016/j.mayocp.2021.07.001",
"article-title": "Acute Kidney Injury in Severe COVID-19 Has Similarities to Sepsis-Associated Kidney Injury: A Multi-Omics Study",
"author": "Alexander",
"doi-asserted-by": "crossref",
"first-page": "2561",
"journal-title": "Mayo Clin. Proc.",
"key": "ref_108",
"volume": "96",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-22781-1",
"article-title": "Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection",
"author": "Diao",
"doi-asserted-by": "crossref",
"first-page": "2506",
"journal-title": "Nat. Commun.",
"key": "ref_109",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/nrneph.2011.16",
"article-title": "Pathophysiology of ischemic acute kidney injury",
"author": "Sharfuddin",
"doi-asserted-by": "crossref",
"first-page": "189",
"journal-title": "Nat. Rev. Nephrol.",
"key": "ref_110",
"volume": "7",
"year": "2011"
},
{
"DOI": "10.3390/biom11081144",
"doi-asserted-by": "crossref",
"key": "ref_111",
"unstructured": "Aranda-Rivera, A.K., Cruz-Gregorio, A., Aparicio-Trejo, O.E., and Pedraza-Chaverri, J. (2021). Mitochondrial Redox Signaling and Oxidative Stress in Kidney Diseases. Biomolecules, 11."
},
{
"DOI": "10.1155/2020/1594726",
"doi-asserted-by": "crossref",
"key": "ref_112",
"unstructured": "Guo, L.P., Liu, S.X., Yang, Q., Liu, H.Y., Xu, L.L., Hao, Y.H., and Zhang, X.Q. (2020). Effect of Thymoquinone on Acute Kidney Injury Induced by Sepsis in BALB/c Mice. Biomed. Res. Int., 2020."
},
{
"DOI": "10.1128/JVI.00964-21",
"article-title": "The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus",
"author": "Dong",
"doi-asserted-by": "crossref",
"first-page": "e0096421",
"journal-title": "J. Virol.",
"key": "ref_113",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1038/s41593-020-00758-5",
"article-title": "Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19",
"author": "Meinhardt",
"doi-asserted-by": "crossref",
"first-page": "168",
"journal-title": "Nat. Neurosci.",
"key": "ref_114",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1126/science.abm2052",
"article-title": "Nervous system consequences of COVID-19",
"author": "Spudich",
"doi-asserted-by": "crossref",
"first-page": "267",
"journal-title": "Science",
"key": "ref_115",
"volume": "375",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2200960119",
"article-title": "Morphological, cellular, and molecular basis of brain infection in COVID-19 patients",
"author": "Crunfli",
"doi-asserted-by": "crossref",
"first-page": "e2200960119",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_116",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.3390/molecules27134142",
"doi-asserted-by": "crossref",
"key": "ref_117",
"unstructured": "Dedoni, S., Avdoshina, V., Camoglio, C., Siddi, C., Fratta, W., Scherma, M., and Fadda, P. (2022). K18- and CAG-hACE2 Transgenic Mouse Models and SARS-CoV-2: Implications for Neurodegeneration Research. Molecules, 27."
},
{
"DOI": "10.1038/s41591-022-02001-z",
"article-title": "Long-term neurologic outcomes of COVID-19",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "2406",
"journal-title": "Nat. Med.",
"key": "ref_118",
"volume": "28",
"year": "2022"
},
{
"DOI": "10.1038/d41586-020-02599-5",
"article-title": "How COVID-19 can damage the brain",
"author": "Marshall",
"doi-asserted-by": "crossref",
"first-page": "342",
"journal-title": "Nature",
"key": "ref_119",
"volume": "585",
"year": "2020"
},
{
"DOI": "10.1002/jmv.28959",
"article-title": "Regulation of autophagy by SARS-CoV-2: The multifunctional contributions of ORF3a",
"author": "Shariq",
"doi-asserted-by": "crossref",
"first-page": "e28959",
"journal-title": "J. Med. Virol.",
"key": "ref_120",
"volume": "95",
"year": "2023"
},
{
"DOI": "10.34172/apb.2023.069",
"article-title": "Targeting Viral ORF3a Protein: A New Approach to Mitigate COVID-19 Induced Immune Cell Apoptosis and Associated Respiratory Complications",
"author": "Treeza",
"doi-asserted-by": "crossref",
"first-page": "678",
"journal-title": "Adv. Pharm. Bull.",
"key": "ref_121",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1016/j.intimp.2022.108595",
"article-title": "MCC950 in the treatment of NLRP3-mediated inflammatory diseases: Latest evidence and therapeutic outcomes",
"author": "Bakhshi",
"doi-asserted-by": "crossref",
"first-page": "108595",
"journal-title": "Int. Immunopharmacol.",
"key": "ref_122",
"volume": "106",
"year": "2022"
},
{
"DOI": "10.1002/ptr.8016",
"article-title": "Cinnamaldehyde inhibits cytokine storms induced by the ORF3a protein of SARS-CoV-2 via ROS-elimination in activated T cells",
"author": "Ma",
"doi-asserted-by": "crossref",
"first-page": "6006",
"journal-title": "Phytother. Res.",
"key": "ref_123",
"volume": "37",
"year": "2023"
},
{
"DOI": "10.1007/s12035-022-02998-x",
"article-title": "Glibenclamide Directly Prevents Neuroinflammation by Targeting SUR1-TRPM4-Mediated NLRP3 Inflammasome Activation in Microglia",
"author": "He",
"doi-asserted-by": "crossref",
"first-page": "6590",
"journal-title": "Mol. Neurobiol.",
"key": "ref_124",
"volume": "59",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2022.105066",
"article-title": "The FDA-approved drug Auranofin has a dual inhibitory effect on SARS-CoV-2 entry and NF-kappaB signaling",
"author": "Laplantine",
"doi-asserted-by": "crossref",
"first-page": "105066",
"journal-title": "iScience",
"key": "ref_125",
"volume": "25",
"year": "2022"
},
{
"DOI": "10.1038/s41598-021-92941-2",
"article-title": "Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2",
"author": "Su",
"doi-asserted-by": "crossref",
"first-page": "13464",
"journal-title": "Sci. Rep.",
"key": "ref_126",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.cyto.2021.155428",
"article-title": "Interleukin-6, procalcitonin and neutrophil-to-lymphocyte ratio: Potential immune-inflammatory parameters to identify severe and fatal forms of COVID-19",
"author": "Sayah",
"doi-asserted-by": "crossref",
"first-page": "155428",
"journal-title": "Cytokine",
"key": "ref_127",
"volume": "141",
"year": "2021"
},
{
"DOI": "10.1038/s41591-020-1051-9",
"article-title": "An inflammatory cytokine signature predicts COVID-19 severity and survival",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "1636",
"journal-title": "Nat. Med.",
"key": "ref_128",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1001/jamanetworkopen.2021.29639",
"article-title": "Association Between Tumor Necrosis Factor Inhibitors and the Risk of Hospitalization or Death Among Patients with Immune-Mediated Inflammatory Disease and COVID-19",
"author": "Izadi",
"doi-asserted-by": "crossref",
"first-page": "e2129639",
"journal-title": "JAMA Netw. Open",
"key": "ref_129",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.1097/CCM.0000000000005591",
"article-title": "A Randomized Double-Blinded Placebo Controlled Trial of Clazakizumab for the Treatment of COVID-19 Pneumonia with Hyperinflammation",
"author": "Lonze",
"doi-asserted-by": "crossref",
"first-page": "1348",
"journal-title": "Crit. Care Med.",
"key": "ref_130",
"volume": "50",
"year": "2022"
},
{
"DOI": "10.1172/jci.insight.134081",
"article-title": "The C5a/C5aR2 axis promotes renal inflammation and tissue damage",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "e134081",
"journal-title": "JCI Insight",
"key": "ref_131",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1186/s12974-021-02274-0",
"article-title": "HMGB1 mediates cognitive impairment caused by the NLRP3 inflammasome in the late stage of traumatic brain injury",
"author": "Tan",
"doi-asserted-by": "crossref",
"first-page": "241",
"journal-title": "J. Neuroinflamm.",
"key": "ref_132",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1038/s41598-021-99072-8",
"article-title": "Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2",
"author": "Lebedeva",
"doi-asserted-by": "crossref",
"first-page": "19481",
"journal-title": "Sci. Rep.",
"key": "ref_133",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1021/jp810638n",
"article-title": "Differential biophysical behavior of human telomeric RNA and DNA quadruplex",
"author": "Arora",
"doi-asserted-by": "crossref",
"first-page": "10515",
"journal-title": "J. Phys. Chem. B",
"key": "ref_134",
"volume": "113",
"year": "2009"
},
{
"DOI": "10.1038/s41421-022-00450-x",
"article-title": "RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models",
"author": "Qin",
"doi-asserted-by": "crossref",
"first-page": "86",
"journal-title": "Cell Discov.",
"key": "ref_135",
"volume": "8",
"year": "2022"
},
{
"key": "ref_136",
"unstructured": "Zhang, J.T., Nasr, M., and Zhao, R.Y. (2024). Unpublished data."
},
{
"article-title": "Fission yeast as a HTS platform for molecular probes of HIV-1 Vpr-induced cell death",
"author": "Benko",
"first-page": "151",
"journal-title": "Int. J. High Throughput Screen.",
"key": "ref_137",
"volume": "1",
"year": "2010"
},
{
"DOI": "10.2174/1570162X17666191128102839",
"article-title": "Development of A Fission Yeast Cell-Based Platform for High Throughput Screening of HIV-1 Protease Inhibitors",
"author": "Benko",
"doi-asserted-by": "crossref",
"first-page": "429",
"journal-title": "Curr. HIV Res.",
"key": "ref_138",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1371/journal.pone.0151286",
"doi-asserted-by": "crossref",
"key": "ref_139",
"unstructured": "Benko, Z., Elder, R.T., Li, G., Liang, D., and Zhao, R.Y. (2016). HIV-1 Protease in the Fission Yeast Schizosaccharomyces pombe. PLoS ONE, 11."
},
{
"DOI": "10.1186/s13578-016-0131-5",
"doi-asserted-by": "crossref",
"key": "ref_140",
"unstructured": "Benko, Z., Liang, D., Li, G., Elder, R.T., Sarkar, A., Takayama, J., Ghosh, A.K., and Zhao, R.Y. (2017). A fission yeast cell-based system for multidrug resistant HIV-1 proteases. Cell Biosci., 7."
},
{
"DOI": "10.1186/2045-3701-2-32",
"doi-asserted-by": "crossref",
"key": "ref_141",
"unstructured": "Yang, H., Nkeze, J., and Zhao, R.Y. (2012). Effects of HIV-1 protease on cellular functions and their potential applications in antiretroviral therapy. Cell Biosci., 2."
},
{
"DOI": "10.3390/pathogens10070804",
"doi-asserted-by": "crossref",
"key": "ref_142",
"unstructured": "Zhang, J., Vernon, K., Li, Q., Benko, Z., Amoroso, A., Nasr, M., and Zhao, R.Y. (2021). Single-Agent and Fixed-Dose Combination HIV-1 Protease Inhibitor Drugs in Fission Yeast (Schizosaccharomyces pombe). Pathogens, 10."
},
{
"DOI": "10.3390/pathogens11070804",
"doi-asserted-by": "crossref",
"key": "ref_143",
"unstructured": "Zhang, J., Li, Q., Kawashima, S.A., Nasr, M., Xue, F., and Zhao, R.Y. (2022). Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model. Pathogens, 11."
},
{
"DOI": "10.2741/zhao",
"article-title": "Yeast perspectives on HIV-1 Vpr",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "905",
"journal-title": "Front. Biosci.",
"key": "ref_144",
"volume": "5",
"year": "2000"
},
{
"DOI": "10.1155/2012/549020",
"doi-asserted-by": "crossref",
"key": "ref_145",
"unstructured": "Andreola, M.L., and Litvak, S. (2012). Yeast and the AIDS virus: The odd couple. J. Biomed. Biotechnol., 2012."
},
{
"DOI": "10.1038/s41423-020-0485-9",
"article-title": "The ORF3a protein of SARS-CoV-2 induces apoptosis in cells",
"author": "Ren",
"doi-asserted-by": "crossref",
"first-page": "881",
"journal-title": "Cell Mol. Immunol.",
"key": "ref_146",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1007/s10495-021-01656-2",
"article-title": "Apoptosis induced by SARS-CoV-2: Can we target it?",
"author": "Donia",
"doi-asserted-by": "crossref",
"first-page": "7",
"journal-title": "Apoptosis",
"key": "ref_147",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1038/s41421-021-00268-z",
"article-title": "The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "31",
"journal-title": "Cell Discov.",
"key": "ref_148",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1016/S2666-5247(20)30004-5",
"article-title": "Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study",
"author": "Chu",
"doi-asserted-by": "crossref",
"first-page": "e14",
"journal-title": "Lancet Microbe",
"key": "ref_149",
"volume": "1",
"year": "2020"
},
{
"DOI": "10.1017/S0950268820002599",
"article-title": "ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection",
"author": "Majumdar",
"doi-asserted-by": "crossref",
"first-page": "e262",
"journal-title": "Epidemiol. Infect.",
"key": "ref_150",
"volume": "148",
"year": "2020"
},
{
"DOI": "10.1016/j.ijantimicag.2020.106272",
"article-title": "Different mutations in SARS-CoV-2 associate with severe and mild outcome",
"author": "Nagy",
"doi-asserted-by": "crossref",
"first-page": "106272",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "ref_151",
"volume": "57",
"year": "2021"
},
{
"DOI": "10.20944/preprints202007.0551.v1",
"doi-asserted-by": "crossref",
"key": "ref_152",
"unstructured": "Tzou, P.L., Tao, K., Nouhin, J., Rhee, S.Y., Hu, B.D., Pai, S., Parkin, N., and Shafer, R.W. (2020). Coronavirus Antiviral Research Database (CoV-RDB): An Online Database Designed to Facilitate Comparisons between Candidate Anti-Coronavirus Compounds. Viruses, 12."
},
{
"DOI": "10.1371/journal.pone.0261045",
"doi-asserted-by": "crossref",
"key": "ref_153",
"unstructured": "Tzou, P.L., Tao, K., Pond, S.L.K., and Shafer, R.W. (2022). Coronavirus Resistance Database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons. PLoS ONE, 17."
},
{
"DOI": "10.1128/MRA.00137-21",
"article-title": "In-Frame 12-Nucleotide Deletion within Open Reading Frame 3a in a SARS-CoV-2 Strain Isolated from a Patient Hospitalized with COVID-19",
"author": "Lednicky",
"doi-asserted-by": "crossref",
"first-page": "10",
"journal-title": "Microbiol. Resour. Announc.",
"key": "ref_154",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1038/s41392-021-00718-w",
"article-title": "Dynamic landscape mapping of humoral immunity to SARS-CoV-2 identifies non-structural protein antibodies associated with the survival of critical COVID-19 patients",
"author": "Cheng",
"doi-asserted-by": "crossref",
"first-page": "304",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "ref_155",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1016/j.isci.2021.103353",
"article-title": "The impact of viral mutations on recognition by SARS-CoV-2 specific T-cells",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "103353",
"journal-title": "iScience",
"key": "ref_156",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1038/d41586-022-00057-y",
"doi-asserted-by": "crossref",
"key": "ref_157",
"unstructured": "(2022). COVID is here to stay: Countries must decide how to adapt. Nature, 601, 165."
},
{
"DOI": "10.1038/s41467-023-38201-5",
"article-title": "Resurgence of Omicron BA.2 in SARS-CoV-2 infection-naive Hong Kong",
"author": "Xie",
"doi-asserted-by": "crossref",
"first-page": "2422",
"journal-title": "Nat. Commun.",
"key": "ref_158",
"volume": "14",
"year": "2023"
}
],
"reference-count": 158,
"references-count": 158,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2076-0817/13/1/75"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Infectious Diseases",
"Microbiology (medical)",
"General Immunology and Microbiology",
"Molecular Biology",
"Immunology and Allergy"
],
"subtitle": [],
"title": "SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects",
"type": "journal-article",
"volume": "13"
}