Investigation of Interactions Between the Protein MPro and the Vanadium Complex VO(metf)2∙H2O: A Computational Approach for COVID-19 Treatment
et al., Biophysica, doi:10.3390/biophysica5010004, Jan 2025
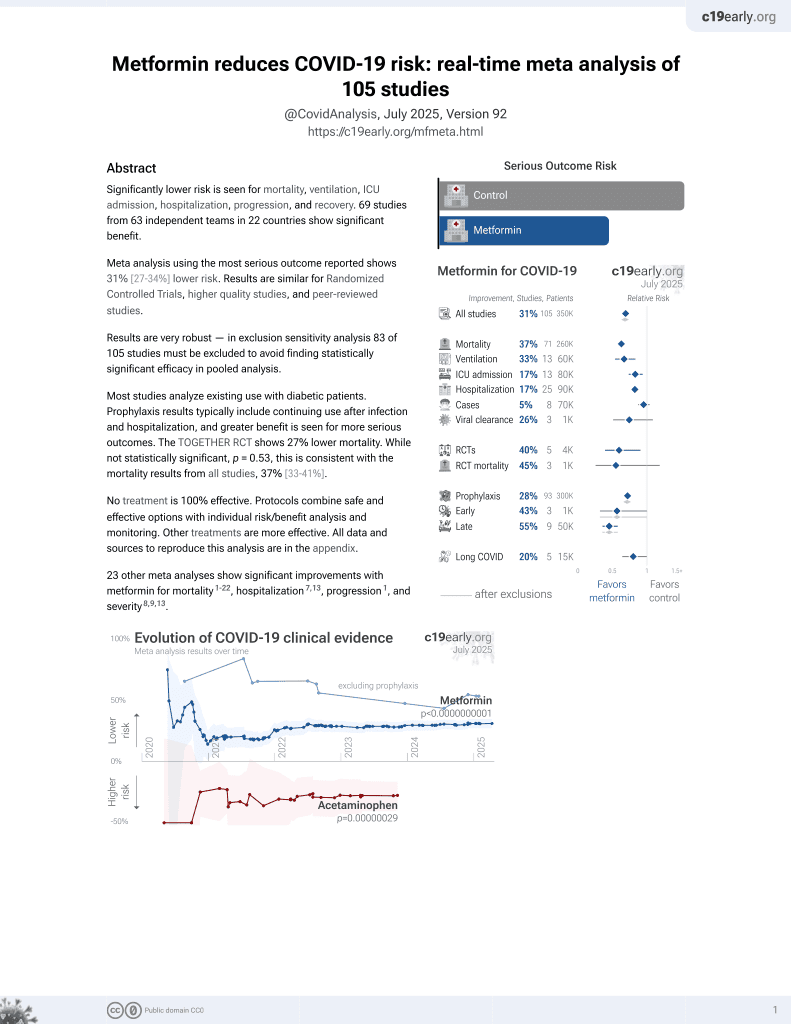
Metformin for COVID-19
3rd treatment shown to reduce risk in
July 2020, now with p < 0.00000000001 from 110 studies.
Lower risk for mortality, ventilation, ICU, hospitalization, progression, recovery, and viral clearance.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico study showing that the vanadium complex VO(metf)2∙H2O (VC) and metformin (MF) may be beneficial for COVID-19 treatment by interacting with the main protease (Mpro) of SARS-CoV-2. Using docking simulations, authors found that VC binds more strongly to Mpro than MF alone, with a binding energy of -126 kcal/mol vs. -75 kcal/mol. Molecular dynamics simulations showed that VC had increased stability and formed more hydrogen bonds with Mpro compared to MF. VC interacted with residues ARG188 and GLU166 of Mpro, while MF interacted with HIE41 and TYR54. The results suggest that complexing metformin with oxovanadium(IV) to form VC may enhance its potential as a COVID-19 treatment by improving its binding affinity and stability with the Mpro active site.
18 preclinical studies support the efficacy of metformin for COVID-19:
A systematic review and meta-analysis of 15 non-COVID-19 preclinical studies showed that metformin inhibits pulmonary inflammation and oxidative stress, minimizes lung injury, and improves survival in animal models of acute respiratory distress syndrome (ARDS) or acute lung injury (ALI)15.
Metformin inhibits SARS-CoV-2 in vitro11,12, minimizes LPS-induced cytokine storm in a mouse model14, minimizes lung damage and fibrosis in a mouse model of LPS-induced ARDS10, may protect against SARS-CoV-2-induced neurological disorders9, may be beneficial via inhibitory effects on ORF3a-mediated inflammasome activation16, reduces UUO and FAN-induced kidney fibrosis10, increases mitochondrial function and decreases TGF-β-induced fibrosis, apoptosis, and inflammation markers in lung epithelial cells10, may reduce inflammation, oxidative stress, and thrombosis via regulating glucose metabolism2, attenuates spike protein S1-induced inflammatory response and α-synuclein aggregation8, may protect against COVID-19 cognitive impairment by suppressing HIF-1α stabilization and reducing neurodegenerative protein aggregation13, may reduce COVID-19 severity and long COVID by inhibiting NETosis via suppression of protein kinase C activation17, enhances interferon responses and reduces SARS-CoV-2 infection and inflammation in diabetic models by suppressing HIF-1α signaling7, may improve COVID-19 outcomes by preventing VDAC1 mistargeting to the plasma membrane, reducing ATP loss, and preserving immune cell function during cytokine storm18, reduces hyperglycemia-induced hepatic ACE2/TMPRSS2 up-regulation and SARS-CoV-2 entry6, may reduce COVID-19 severity by suppressing monocyte inflammatory responses and glycolytic activation via AMPK pathway modulation5, and may improve outcomes via modulation of immune responses with increased anti-inflammatory T lymphocyte gene expression and via enhanced gut microbiota diversity19.
1.
Tavares et al., Investigation of Interactions Between the Protein MPro and the Vanadium Complex VO(metf)2∙H2O: A Computational Approach for COVID-19 Treatment, Biophysica, doi:10.3390/biophysica5010004.
2.
Hou et al., Metformin is a potential therapeutic for COVID-19/LUAD by regulating glucose metabolism, Scientific Reports, doi:10.1038/s41598-024-63081-0.
3.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
4.
Lockwood, T., Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent, BioMetals, doi:10.1007/s10534-024-00590-5.
5.
Maurmann et al., Immunoregulatory effect of metformin in monocytes exposed to SARS-CoV-2 spike protein subunit 1, bioRxiv, doi:10.1101/2025.09.12.675877.
6.
Rao et al., Pathological Glucose Levels Enhance Entry Factor Expression and Hepatic SARS‐CoV‐2 Infection, Journal of Cellular and Molecular Medicine, doi:10.1111/jcmm.70581.
7.
Joshi et al., Severe SARS‐CoV‐2 infection in diabetes was rescued in mice supplemented with metformin and/or αKG, and patients taking metformin, via HIF1α‐IFN axis, Clinical and Translational Medicine, doi:10.1002/ctm2.70275.
8.
Chang et al., SARS-CoV-2 Spike Protein 1 Causes Aggregation of α-Synuclein via Microglia-Induced Inflammation and Production of Mitochondrial ROS: Potential Therapeutic Applications of Metformin, Biomedicines, doi:10.3390/biomedicines12061223.
9.
Yang et al., SARS-CoV-2 infection causes dopaminergic neuron senescence, Cell Stem Cell, doi:10.1016/j.stem.2023.12.012.
10.
Miguel et al., Enhanced fatty acid oxidation through metformin and baicalin as therapy for COVID-19 and associated inflammatory states in lung and kidney, Redox Biology, doi:10.1016/j.redox.2023.102957.
11.
Ventura-López et al., Treatment with metformin glycinate reduces SARS-CoV-2 viral load: An in vitro model and randomized, double-blind, Phase IIb clinical trial, Biomedicine & Pharmacotherapy, doi:10.1016/j.biopha.2022.113223.
12.
Parthasarathy et al., Metformin Suppresses SARS-CoV-2 in Cell Culture, bioRxiv, doi:10.1101/2021.11.18.469078.
13.
Lee et al., SARS-CoV-2 spike protein causes synaptic dysfunction and p-tau and α-synuclein aggregation leading cognitive impairment: The protective role of metformin, PLOS One, doi:10.1371/journal.pone.0336015.
14.
Taher et al., Anti‑inflammatory effect of metformin against an experimental model of LPS‑induced cytokine storm, Experimental and Therapeutic Medicine, doi:10.3892/etm.2023.12114.
15.
Wang et al., Effects of metformin on acute respiratory distress syndrome in preclinical studies: a systematic review and meta-analysis, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1215307.
16.
Zhang et al., SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects, Pathogens, doi:10.3390/pathogens13010075.
17.
Monsalve et al., NETosis: A key player in autoimmunity, COVID-19, and long COVID, Journal of Translational Autoimmunity, doi:10.1016/j.jtauto.2025.100280.
Tavares et al., 31 Jan 2025, Brazil, peer-reviewed, 5 authors.
Contact: teo@ufla.br (corresponding author), tavares.camila@outlook.com, eduardo.benedito1@estudante.ufla.br, tainah-martins@hotmail.com, rodrigomancini4@gmail.com, vc@n3.e.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Investigation of Interactions Between the Protein MPro and the Vanadium Complex VO(metf)2∙H2O: A Computational Approach for COVID-19 Treatment
Biophysica, doi:10.3390/biophysica5010004
Since 2020, the attention of the scientific community has been focused on the overwhelming COVID-19 pandemic, the infectious disease caused by the coronavirus that has affected populations worldwide. The alarming number of deaths and the severity of the symptoms have driven studies aimed at combating this disease. One of the key components in the development of this disease is the protein M Pro , responsible for the replication and transcription of the virus, making it an excellent biological target in research efforts seeking an effective treatment for the disease. Furthermore, studies have shown that vanadium complexes, such as bis(N ′ ,N ′ -dimethylbiguanide)oxovanadium (IV), VO(metf) 2 •H 2 O, exhibit highly promising effects for the treatment of COVID-19. This molecule contains a ligand known as metformin, which also holds a prominent place as a potential agent in the treatment of this disease due to its antiviral properties. Therefore, an investigation into the interactions between these two systems (M Pro +Vanadium Complex and M Pro +Metformin) is pertinent given the significance of these two molecules. Thus, computational studies such as molecular docking and classical molecular dynamics are considered advantageous, assisting in this comparative study, as well as providing a deeper understanding of the interactions that occur within each of them.
References
Anghebem, Rego, Picheth, COVID-19 e Diabetes: A relação entre duas pandemias distintas, Rev. Bras. Anal. Clin
Badran, Hassan, Manasrah, Nassar, Experimental and theoretical studies on the thermal decomposition of metformin, J. Therm. Anal. Calorim, doi:10.1007/s10973-019-08213-9
Beshbishy, Oti, Hussein, Rehan, Adeyemi et al., Factors behind the higher COVID-19 risk in diabetes: A critical review, Front. Public Health, doi:10.3389/fpubh.2021.591982
Biovia, Discovery Studio Visualizer
Cárdenas, Marquetand, Mai, González, A force field for a manganese-vanadium water oxidation catalyst: Redox potentials in solution as showcase, Catalysts, doi:10.3390/catal11040493
De Vivo, Masetti, Bottegoni, Cavalli, Role of molecular dynamics and related methods in drug discovery, J. Med. Chem, doi:10.1021/acs.jmedchem.5b01684
Dolezal, Soukup, Malinak, Savedra, Marek et al., Towards understanding the mechanism of action of antibacterial N-alkyl-3-hydroxypyridinium salts: Biological activities, molecular modeling and QSAR studies, Eur. J. Med. Chem, doi:10.1016/j.ejmech.2016.05.058
El-Arabey, Abdalla, Abd-Allah, Marenga, Modafer et al., Molecular dynamic and bioinformatic studies of metformin-induced ACE2 phosphorylation in the presence of different SARS-CoV-2 S protein mutations, Saudi J. Biol. Sci, doi:10.1016/j.sjbs.2023.103699
El-Arabey, Zhang, Abdalla, Al-Shouli, Alkhalil et al., Metformin as a promising target for DPP4 expression: Computational modeling and experimental validation, Med. Oncol, doi:10.1007/s12032-023-02140-4
Esakandari, Nabi-Afjadi, Fakkari-Afjadi, Farahmandian, Miresmaeili et al., A comprehensive review of COVID-19 characteristics, Biol. Proced. Online, doi:10.1186/s12575-020-00128-2
França, Pascutti, Ramalho, Figueroa-Villar, A three-dimensional structure of Plasmodium falciparum serine hydroxymethyltransferase in complex with glycine and 5-formyl-tetrahydrofolate. Homology modeling and molecular dynamics, Biophys. Chem, doi:10.1016/j.bpc.2004.12.002
Gasmi, Peana, Pivina, Srinath, Benahmed et al., Interrelations between COVID-19 and other disorders, Clin. Immunol, doi:10.1016/j.clim.2020.108651
Giacoppo, França, Kuča, Da Cunha, Abagyan et al., Molecular modeling and in vitro reactivation study between the oxime BI-6 and acetylcholinesterase inhibited by different nerve agents, J Biomol. Struct. Dyn, doi:10.1080/07391102.2014.989408
Halim, Waqas, Khan, Al-Harrasi, In silico prediction of novel inhibitors of SARS-CoV-2 main protease through structure-based virtual screening and molecular dynamic simulation, Pharmaceuticals, doi:10.3390/ph14090896
Hariyanto, Kurniawan, Metformin use is associated with reduced mortality rate from coronavirus disease 2019 (COVID-19) infection, Obes. Med, doi:10.1016/j.obmed.2020.100290
Humphrey, Dalke, Schulten, Vmd, Visual molecular dynamics, J. Mol. Graph
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kazakou, Lambadiari, Ikonomidis, Kountouri, Panagopoulos et al., Diabetes and COVID-19; A Bidirectional Interplay, Front. Endocrinol, doi:10.3389/fendo.2022.780663
Lin, Li, Lin, A review on applications of computational methods in drug screening and design, Molecules, doi:10.3390/molecules25061375
Magalhães, Barbosa, Dardenne, Métodos de docking receptor-ligante para o desenho racional de compostos bioativos
Martínez, Borin, Skaf, Fundamentos de simulação por dinâmica molecular
Morgon, Coutinho, Livraria da Física
Morgon, Coutinho, Livraria da Física
Neese, The ORCA program system, Wiley Interdiscip. Rev. Comput. Mol. Sci, doi:10.1002/wcms.81
Pandey, Fernandez, Gentile, Isayev, Tropsha et al., The transformational role of GPU computing and deep learning in drug discovery, Nat. Mach. Intell, doi:10.1038/s42256-022-00463-x
Qu, Zheng, Wang, Li, Liu et al., The potential effects of clinical antidiabetic agents on SARS-CoV-2, J. Diabetes, doi:10.1111/1753-0407.13135
Ramalho, De Alencastro, La-Scalea, Figueroa-Villar, Theoretical evaluation of adiabatic and vertical electron affinity of some radiosensitizers in solution using FEP, ab initio and DFT methods, Biophys. Chem, doi:10.1016/j.bpc.2004.03.002
Rashedi, Mahdavi Poor, Asgharzadeh, Pourostadi, Samadi Kafil et al., Risk factors for COVID-19, Infez. Med
Renganayaki, Srinivasan, Hf, DFT computations and spectroscopic study of the vibrational and thermodynamic properties of metformin, Int. J. Pharmtech Res
Rocha, Analysis of Biocide Action from Organotin Compounds: Insights from Theoric and Experimental Models
Rusanov, Zou, Babak, Biological properties of transition metal complexes with metformin and its analogues, Pharmaceuticals, doi:10.3390/ph15040453
Schrödinger, The PyMOL Molecular Graphics System
Scior, Abdallah, Mustafa, Guevara-García, Rehder, Are vanadium complexes druggable against the main protease mpro of SARS-CoV-2?-A computational approach, Inorganica Chim. Acta, doi:10.1016/j.ica.2021.120287
Semiz, Vanadium as potential therapeutic agent for COVID-19: A focus on its antiviral, antiinflamatory, and antihyperglycemic effects, J. Trace Elements Med. Biol, doi:10.1016/j.jtemb.2021.126887
Sharma, Deep, In-silico drug repurposing for targeting SARS-CoV-2 main protease (Mpro), J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1844058
Solerte, Trevisan, Lovati, Rossi, Pastore et al., Sitagliptin Treatment at the Time of Hospitalization Was Associated With Reduced Mortality in Patients With Type 2 Diabetes and COVID-19: A Multicenter, Case-Control, Retrospective, Observational Study, Diabetes Care, doi:10.2337/dc20-1521
Tavares, Santos, Da Cunha, Ramalho, Molecular dynamics-assisted interaction of vanadium complex-AMPK: From force field development to biological application for Alzheimer's treatment, J. Phys. Chem. B, doi:10.1021/acs.jpcb.2c07147
Thomsen, Christensen, Moldock, A new technique for high-accuracy molecular docking, J. Med. Chem, doi:10.1021/jm051197e
Torres, Sodero, Jofily, Silva-Jr, Key topics in molecular docking for drug design, Int. J. Mol. Sci, doi:10.3390/ijms20184574
Xue, Yu, Yang, Xue, Wu et al., Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design, J. Virol, doi:10.1128/JVI.02114-07
Yang, Xie, Xue, Yang, Ma et al., Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases, PLoS Biol, doi:10.1371/journal.pbio.0030324
DOI record:
{
"DOI": "10.3390/biophysica5010004",
"ISSN": [
"2673-4125"
],
"URL": "http://dx.doi.org/10.3390/biophysica5010004",
"abstract": "<jats:p>Since 2020, the attention of the scientific community has been focused on the overwhelming COVID-19 pandemic, the infectious disease caused by the coronavirus that has affected populations worldwide. The alarming number of deaths and the severity of the symptoms have driven studies aimed at combating this disease. One of the key components in the development of this disease is the protein MPro, responsible for the replication and transcription of the virus, making it an excellent biological target in research efforts seeking an effective treatment for the disease. Furthermore, studies have shown that vanadium complexes, such as bis(N′,N′-dimethylbiguanide)oxovanadium (IV), VO(metf)2∙H2O, exhibit highly promising effects for the treatment of COVID-19. This molecule contains a ligand known as metformin, which also holds a prominent place as a potential agent in the treatment of this disease due to its antiviral properties. Therefore, an investigation into the interactions between these two systems (MPro+Vanadium Complex and MPro+Metformin) is pertinent given the significance of these two molecules. Thus, computational studies such as molecular docking and classical molecular dynamics are considered advantageous, assisting in this comparative study, as well as providing a deeper understanding of the interactions that occur within each of them.</jats:p>",
"alternative-id": [
"biophysica5010004"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-0710-3956",
"affiliation": [
{
"name": "Institute of Chemistry, University of Campinas, Campinas 13083-970, SP, Brazil"
}
],
"authenticated-orcid": false,
"family": "Tavares",
"given": "Camila A.",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Laboratory of Molecular Modelling, Department of Chemistry, Federal University of Lavras, Lavras 37200-000, MG, Brazil"
}
],
"family": "Benedito",
"given": "Eduardo F.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Molecular Modelling, Department of Chemistry, Federal University of Lavras, Lavras 37200-000, MG, Brazil"
}
],
"family": "Santos",
"given": "Taináh M. R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Molecular Modelling, Department of Chemistry, Federal University of Lavras, Lavras 37200-000, MG, Brazil"
}
],
"family": "Santos",
"given": "Rodrigo M.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7324-1353",
"affiliation": [
{
"name": "Laboratory of Molecular Modelling, Department of Chemistry, Federal University of Lavras, Lavras 37200-000, MG, Brazil"
},
{
"name": "Department of Chemistry, Faculty of Science, University of Hradec Králové, 500 03 Hradec Králové, Czech Republic"
}
],
"authenticated-orcid": false,
"family": "Ramalho",
"given": "Teodorico C.",
"sequence": "additional"
}
],
"container-title": "Biophysica",
"container-title-short": "Biophysica",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
1,
31
]
],
"date-time": "2025-01-31T10:08:26Z",
"timestamp": 1738318106000
},
"deposited": {
"date-parts": [
[
2025,
1,
31
]
],
"date-time": "2025-01-31T10:37:19Z",
"timestamp": 1738319839000
},
"funder": [
{
"name": "Brazilian financial agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico"
},
{
"name": "Fundação de Amparo ao Ensino e Pesquisa de Minas Gerais"
},
{
"DOI": "10.13039/501100002322",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100002322",
"id-type": "DOI"
}
],
"name": "Coordenação de Aperfeiçoamento de Pessoal de Nível Superior"
},
{
"name": "Federal University of Lavras (UFLA)"
},
{
"award": [
"VT2019–2021"
],
"name": "Long-term development plan University of Hradec Králové"
}
],
"indexed": {
"date-parts": [
[
2025,
2,
1
]
],
"date-time": "2025-02-01T05:21:12Z",
"timestamp": 1738387272783,
"version": "3.35.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
1,
31
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2025,
3
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
1,
31
]
],
"date-time": "2025-01-31T00:00:00Z",
"timestamp": 1738281600000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2673-4125/5/1/4/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "4",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
1,
31
]
]
},
"published-online": {
"date-parts": [
[
2025,
1,
31
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1186/s12575-020-00128-2",
"doi-asserted-by": "crossref",
"key": "ref_1",
"unstructured": "Esakandari, H., Nabi-Afjadi, M., Fakkari-Afjadi, J., Farahmandian, N., Miresmaeili, S.-M., and Bahreini, E. (2020). A comprehensive review of COVID-19 characteristics. Biol. Proced. Online, 22."
},
{
"article-title": "Risk factors for COVID-19",
"author": "Rashedi",
"first-page": "469",
"journal-title": "Infez. Med.",
"key": "ref_2",
"volume": "28",
"year": "2020"
},
{
"key": "ref_3",
"unstructured": "(2024, December 31). WHO COVID-19 Dashboard. Available online: https://covid19.who.int/."
},
{
"article-title": "COVID-19 e Diabetes: A relação entre duas pandemias distintas",
"author": "Anghebem",
"first-page": "154",
"journal-title": "Rev. Bras. Anal. Clin.",
"key": "ref_4",
"volume": "52",
"year": "2020"
},
{
"DOI": "10.1016/j.clim.2020.108651",
"article-title": "Interrelations between COVID-19 and other disorders",
"author": "Gasmi",
"doi-asserted-by": "crossref",
"first-page": "108651",
"journal-title": "Clin. Immunol.",
"key": "ref_5",
"volume": "224",
"year": "2021"
},
{
"DOI": "10.3389/fpubh.2021.591982",
"doi-asserted-by": "crossref",
"key": "ref_6",
"unstructured": "Beshbishy, A.M., Oti, V.B., Hussein, D.E., Rehan, I.F., Adeyemi, O.S., Rivero-Perez, N., Zaragoza-Bastida, A., Shah, M.A., Abouelezz, K., and Hetta, H.F. (2021). Factors behind the higher COVID-19 risk in diabetes: A critical review. Front. Public Health, 9."
},
{
"DOI": "10.3389/fendo.2022.780663",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Kazakou, P., Lambadiari, V., Ikonomidis, I., Kountouri, A., Panagopoulos, G., Athanasopoulos, S., Korompoki, E., Kalomenidis, I., Dimopoulos, M.A., and Mitrakou, A. (2022). Diabetes and COVID-19; A Bidirectional Interplay. Front. Endocrinol., 13."
},
{
"DOI": "10.1016/j.jtemb.2021.126887",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Semiz, S. (2022). Vanadium as potential therapeutic agent for COVID-19: A focus on its antiviral, antiinflamatory, and antihyperglycemic effects. J. Trace Elements Med. Biol., 69."
},
{
"DOI": "10.1111/1753-0407.13135",
"article-title": "The potential effects of clinical antidiabetic agents on SARS-CoV-2",
"author": "Qu",
"doi-asserted-by": "crossref",
"first-page": "243",
"journal-title": "J. Diabetes",
"key": "ref_9",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1016/j.sjbs.2023.103699",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "El-Arabey, A.A., Abdalla, M., Abd-Allah, A.R., Marenga, H.S., Modafer, Y., and Aloufi, A.S. (2023). Molecular dynamic and bioinformatic studies of metformin-induced ACE2 phosphorylation in the presence of different SARS-CoV-2 S protein mutations. Saudi J. Biol. Sci., 30."
},
{
"DOI": "10.1016/j.obmed.2020.100290",
"article-title": "Metformin use is associated with reduced mortality rate from coronavirus disease 2019 (COVID-19) infection",
"author": "Hariyanto",
"doi-asserted-by": "crossref",
"first-page": "100290",
"journal-title": "Obes. Med.",
"key": "ref_11",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.2337/dc20-1521",
"article-title": "Sitagliptin Treatment at the Time of Hospitalization Was Associated With Reduced Mortality in Patients With Type 2 Diabetes and COVID-19: A Multicenter, Case-Control, Retrospective, Observational Study",
"author": "Solerte",
"doi-asserted-by": "crossref",
"first-page": "2999",
"journal-title": "Diabetes Care",
"key": "ref_12",
"volume": "43",
"year": "2020"
},
{
"DOI": "10.1371/journal.pbio.0030428",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Yang, H., Xie, W., Xue, X., Yang, K., Ma, J., Liang, W., Zhao, Q., Zhou, Z., Pei, D., and Ziebuhr, J. (2005). Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases. PLoS Biol., 3."
},
{
"DOI": "10.1128/JVI.02114-07",
"article-title": "Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design",
"author": "Xue",
"doi-asserted-by": "crossref",
"first-page": "2515",
"journal-title": "J. Virol.",
"key": "ref_14",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.3390/ph15040453",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Rusanov, D.A., Zou, J., and Babak, M.V. (2022). Biological properties of transition metal complexes with metformin and its analogues. Pharmaceuticals, 15."
},
{
"DOI": "10.1016/j.ica.2021.120287",
"article-title": "Are vanadium complexes druggable against the main protease mpro of SARS-CoV-2?—A computational approach",
"author": "Scior",
"doi-asserted-by": "crossref",
"first-page": "120287",
"journal-title": "Inorganica Chim. Acta",
"key": "ref_16",
"volume": "519",
"year": "2021"
},
{
"DOI": "10.3390/molecules25061375",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Lin, X., Li, X., and Lin, X. (2020). A review on applications of computational methods in drug screening and design. Molecules, 25."
},
{
"DOI": "10.1016/j.ejmech.2016.05.058",
"article-title": "Towards understanding the mechanism of action of antibacterial N-alkyl-3-hydroxypyridinium salts: Biological activities, molecular modeling and QSAR studies",
"author": "Dolezal",
"doi-asserted-by": "crossref",
"first-page": "699",
"journal-title": "Eur. J. Med. Chem.",
"key": "ref_18",
"volume": "121",
"year": "2016"
},
{
"DOI": "10.1016/j.bpc.2004.12.002",
"article-title": "A three-dimensional structure of Plasmodium falciparum serine hydroxymethyltransferase in complex with glycine and 5-formyl-tetrahydrofolate. Homology modeling and molecular dynamics",
"author": "Pascutti",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Biophys. Chem.",
"key": "ref_19",
"volume": "115",
"year": "2005"
},
{
"DOI": "10.1016/j.bpc.2004.03.002",
"article-title": "Theoretical evaluation of adiabatic and vertical electron affinity of some radiosensitizers in solution using FEP, ab initio and DFT methods",
"author": "Ramalho",
"doi-asserted-by": "crossref",
"first-page": "267",
"journal-title": "Biophys. Chem.",
"key": "ref_20",
"volume": "110",
"year": "2004"
},
{
"DOI": "10.1080/07391102.2014.989408",
"article-title": "Molecular modeling and in vitro reactivation study between the oxime BI-6 and acetylcholinesterase inhibited by different nerve agents",
"author": "Giacoppo",
"doi-asserted-by": "crossref",
"first-page": "2048",
"journal-title": "J Biomol. Struct. Dyn.",
"key": "ref_21",
"volume": "9",
"year": "2015"
},
{
"DOI": "10.3390/ijms20184574",
"doi-asserted-by": "crossref",
"key": "ref_22",
"unstructured": "Torres, P.H., Sodero, A.C., Jofily, P., and Silva-Jr, F.P. (2019). Key topics in molecular docking for drug design. Int. J. Mol. Sci., 20."
},
{
"key": "ref_23",
"unstructured": "Morgon, N.H., and Coutinho, K. (2007). Métodos de docking receptor-ligante para o desenho racional de compostos bioativos. Métodos de Química Teórica e Modelagem Molecular, Livraria da Física. [1st ed.]."
},
{
"key": "ref_24",
"unstructured": "Rocha, M.V.J. (2010). Analysis of Biocide Action from Organotin Compounds: Insights from Theoric and Experimental Models. [Master’s Thesis, Federal University of Lavras]."
},
{
"DOI": "10.1021/acs.jmedchem.5b01684",
"article-title": "Role of molecular dynamics and related methods in drug discovery",
"author": "Masetti",
"doi-asserted-by": "crossref",
"first-page": "4035",
"journal-title": "J. Med. Chem.",
"key": "ref_25",
"volume": "59",
"year": "2016"
},
{
"key": "ref_26",
"unstructured": "Morgon, N.H., and Coutinho, K. (2007). Fundamentos de simulação por dinâmica molecular. Métodos de Química Teórica e Modelagem Molecular, Livraria da Física. [1st ed.]."
},
{
"DOI": "10.1007/s10973-019-08213-9",
"article-title": "Experimental and theoretical studies on the thermal decomposition of metformin",
"author": "Badran",
"doi-asserted-by": "crossref",
"first-page": "433",
"journal-title": "J. Therm. Anal. Calorim.",
"key": "ref_27",
"volume": "138",
"year": "2019"
},
{
"article-title": "HF, DFT computations and spectroscopic study of the vibrational and thermodynamic properties of metformin",
"author": "Renganayaki",
"first-page": "1350",
"journal-title": "Int. J. Pharmtech Res.",
"key": "ref_28",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.1021/acs.jpcb.2c07147",
"article-title": "Molecular dynamics-assisted interaction of vanadium complex–AMPK: From force field development to biological application for Alzheimer’s treatment",
"author": "Tavares",
"doi-asserted-by": "crossref",
"first-page": "495",
"journal-title": "J. Phys. Chem. B",
"key": "ref_29",
"volume": "127",
"year": "2023"
},
{
"DOI": "10.3390/catal11040493",
"doi-asserted-by": "crossref",
"key": "ref_30",
"unstructured": "Cárdenas, G., Marquetand, P., Mai, S., and González, L. (2021). A force field for a manganese-vanadium water oxidation catalyst: Redox potentials in solution as showcase. Catalysts, 11."
},
{
"DOI": "10.1002/wcms.81",
"article-title": "The ORCA program system",
"author": "Neese",
"doi-asserted-by": "crossref",
"first-page": "73",
"journal-title": "Wiley Interdiscip. Rev. Comput. Mol. Sci.",
"key": "ref_31",
"volume": "2",
"year": "2012"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors",
"author": "Jin",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "ref_32",
"volume": "582",
"year": "2020"
},
{
"key": "ref_33",
"unstructured": "BIOVIA (2021). Discovery Studio Visualizer, Dassault Systemes."
},
{
"DOI": "10.1021/jm051197e",
"article-title": "MolDock: A new technique for high-accuracy molecular docking",
"author": "Thomsen",
"doi-asserted-by": "crossref",
"first-page": "3315",
"journal-title": "J. Med. Chem.",
"key": "ref_34",
"volume": "49",
"year": "2006"
},
{
"DOI": "10.1038/s42256-022-00463-x",
"article-title": "The transformational role of GPU computing and deep learning in drug discovery",
"author": "Pandey",
"doi-asserted-by": "crossref",
"first-page": "211",
"journal-title": "Nat. Mach. Intell.",
"key": "ref_35",
"volume": "4",
"year": "2022"
},
{
"key": "ref_36",
"unstructured": "(2020). AMBER 2020, University of California."
},
{
"DOI": "10.3390/ph14090896",
"doi-asserted-by": "crossref",
"key": "ref_37",
"unstructured": "Halim, S.A., Waqas, M., Khan, A., and Al-Harrasi, A. (2021). In silico prediction of novel inhibitors of SARS-CoV-2 main protease through structure-based virtual screening and molecular dynamic simulation. Pharmaceuticals, 14."
},
{
"DOI": "10.1080/07391102.2020.1844058",
"article-title": "In-silico drug repurposing for targeting SARS-CoV-2 main protease (Mpro)",
"author": "Sharma",
"doi-asserted-by": "crossref",
"first-page": "3003",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "ref_38",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1007/s12032-023-02140-4",
"article-title": "Metformin as a promising target for DPP4 expression: Computational modeling and experimental validation",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Med. Oncol.",
"key": "ref_39",
"volume": "40",
"year": "2023"
},
{
"key": "ref_40",
"unstructured": "Schrödinger, LLC (2015). The PyMOL Molecular Graphics System, Schrödinger, LLC."
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"article-title": "VMD: Visual molecular dynamics",
"author": "Humphrey",
"doi-asserted-by": "crossref",
"first-page": "33",
"journal-title": "J. Mol. Graph.",
"key": "ref_41",
"volume": "14",
"year": "1996"
}
],
"reference-count": 41,
"references-count": 41,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2673-4125/5/1/4"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Investigation of Interactions Between the Protein MPro and the Vanadium Complex VO(metf)2∙H2O: A Computational Approach for COVID-19 Treatment",
"type": "journal-article",
"volume": "5"
}