Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2
et al., Scientific Reports, doi:10.1038/s41598-024-58532-7, Apr 2024
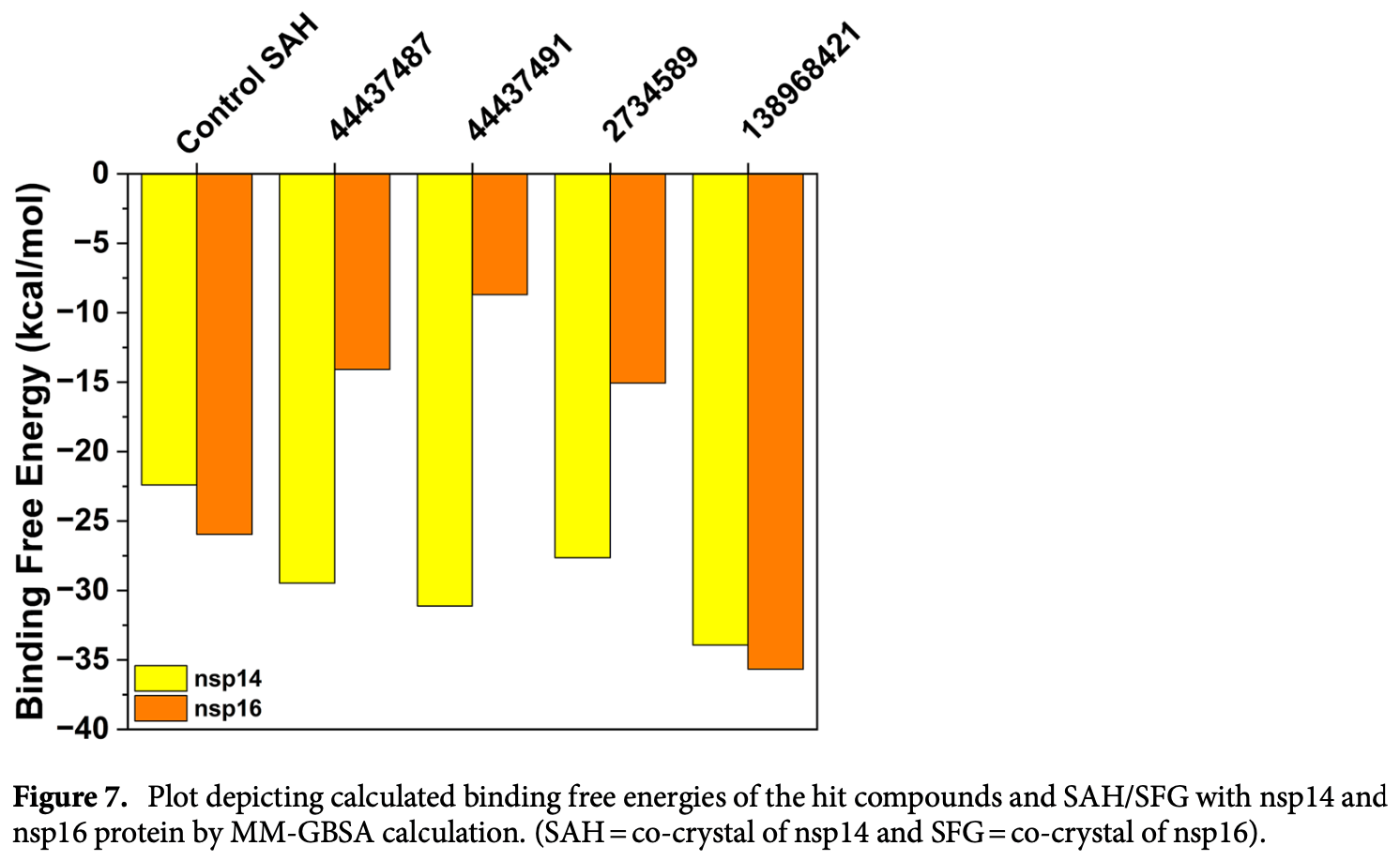
In silico study showing that andrographolide derivatives (PubChem CID 2734589 and 138968421) are potential dual inhibitors of SARS-CoV-2 methyltransferases nsp14 and nsp16, which are crucial for viral replication and evading host immune responses. Authors conducted virtual screening, molecular docking, ADMET profiling, molecular dynamics simulations, free-energy landscape analysis, and density functional theory calculations on 413 andrographolide derivatives. The two lead compounds formed stable complexes with nsp14 and nsp16 proteins via hydrophobic interactions, hydrogen bonds and electrostatic interactions, and had predicted class four toxicity with LD50 values of 500-700 mg/kg. Molecular dynamics simulations and binding free energy calculations confirmed the stability of the protein-ligand complexes.
25 preclinical studies support the efficacy of andrographolide for COVID-19:
In vitro studies demonstrate inhibition of the MproA,18 protein.
In vitro studies demonstrate efficacy in Calu-3B,18, A549C,14, and HUVECD,18 cells.
Animal studies demonstrate efficacy in Sprague Dawley miceE,18 and Golden Syrian hamstersF,14.
Andrographolide inhibits Mpro in a dose-dependent manner18, reduces ACE2 levels in the lung tissue of mice in combination with baicalein18, inhibits binding between the SARS-CoV-2 spike protein and ACE218, alleviates lung inflammation and cytokine storm in mice18, and improves survival and reduces lung inflammation via anti-inflammatory effects in Syrian hamsters14.
1.
Zhang et al., Effects and Mechanisms of Andrographolide for COVID-19: A Network Pharmacology-Based and Experimentally Validated Study, Natural Product Communications, doi:10.1177/1934578X241288428.
2.
Thomas et al., Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-58532-7.
3.
Arifin et al., Computational exploration of Andrographis paniculata herb compounds as potential antiviral agents targeting NSP3 (6W02) and NSP5 (7AR6) of SARS-COV-2, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2023.25.2.0292.
4.
Bhattarai et al., Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2174596.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
6.
Dassanayake et al., Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus, International Journal of Molecular Sciences, doi:10.3390/ijms231911131.
7.
Ningrum et al., Potency Of Andrographolide, L-Mimosine And Asiaticoside Compound As Antiviral For Covid-19 Based On In Silico Method, Proceedings Universitas Muhammadiyah Yogyakarta Undergraduate Conference, doi:10.18196/umygrace.v2i2.418.
8.
Ravichandran et al., Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach, Biointerface Research in Applied Chemistry, doi:10.33263/BRIAC132.155.
9.
Saeheng et al., In Silico Prediction of Andrographolide Dosage Regimens for COVID-19 Treatment, The American Journal of Chinese Medicine, doi:10.1142/S0192415X22500732.
10.
Khanal et al., Combination of system biology to probe the anti-viral activity of andrographolide and its derivative against COVID-19, RSC Advances, doi:10.1039/D0RA10529E.
11.
Rehan et al., A Computational Approach Identified Andrographolide as a Potential Drug for Suppressing COVID-19-Induced Cytokine Storm, Frontiers in Immunology, doi:10.3389/fimmu.2021.648250.
12.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
13.
Dey et al., The role of andrographolide and its derivative in COVID-19 associated proteins and immune system, Research Square, doi:10.21203/rs.3.rs-35800/v1.
14.
Kongsomros et al., In vivo evaluation of Andrographis paniculata and Boesenbergia rotunda extract activity against SARS-CoV-2 Delta variant in Golden Syrian hamsters: Potential herbal alternative for COVID-19 treatment, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2024.05.004.
15.
Chaopreecha et al., Andrographolide attenuates SARS-CoV-2 infection via an up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC), Phytomedicine, doi:10.1016/j.phymed.2024.156279.
16.
Li et al., Andrographolide suppresses SARS-CoV-2 infection by downregulating ACE2 expression: A mechanistic study, Antiviral Therapy, doi:10.1177/13596535241259952.
17.
Low et al., The wide spectrum anti-inflammatory activity of andrographolide in comparison to NSAIDs: a promising therapeutic compound against the cytokine storm, bioRxiv, doi:10.1101/2024.02.21.581396.
18.
Wan et al., Synergistic inhibition effects of andrographolide and baicalin on coronavirus mechanisms by downregulation of ACE2 protein level, Scientific Reports, doi:10.1038/s41598-024-54722-5.
19.
Siridechakorn et al., Inhibitory efficiency of Andrographis paniculata extract on viral multiplication and nitric oxide production, Scientific Reports, doi:10.1038/s41598-023-46249-y.
a.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
b.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
c.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
d.
HUVEC (Human Umbilical Vein Endothelial Cells) are primary endothelial cells derived from the vein of the umbilical cord. They are used to study vascular biology, including inflammation, angiogenesis, and viral interactions with endothelial cells.
e.
An outbred multipurpose breed of albino mouse used extensively in medical research.
f.
A rodent model widely used in infectious disease research due to their susceptibility to viral infections and similar disease progression to humans.
Thomas et al., 29 Apr 2024, peer-reviewed, 4 authors.
Contact: jsatija11@gmail.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2
Scientific Reports, doi:10.1038/s41598-024-58532-7
The Covid-19 pandemic outbreak has accelerated tremendous efforts to discover a therapeutic strategy that targets severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to control viral infection. Various viral proteins have been identified as potential drug targets, however, to date, no specific therapeutic cure is available against the SARS-CoV-2. To address this issue, the present work reports a systematic cheminformatic approach to identify the potent andrographolide derivatives that can target methyltransferases of SARS-CoV-2, i.e. nsp14 and nsp16 which are crucial for the replication of the virus and host immune evasion. A consensus of cheminformatics methodologies including virtual screening, molecular docking, ADMET profiling, molecular dynamics simulations, free-energy landscape analysis, molecular mechanics generalized born surface area (MM-GBSA), and density functional theory (DFT) was utilized. Our study reveals two new andrographolide derivatives (PubChem CID: 2734589 and 138968421) as natural bioactive molecules that can form stable complexes with both proteins via hydrophobic interactions, hydrogen bonds and electrostatic interactions. The toxicity analysis predicts class four toxicity for both compounds with LD 50 value in the range of 500-700 mg/kg. MD simulation reveals the stable formation of the complex for both the compounds and their average trajectory values were found to be lower than the control inhibitor and protein alone. MMGBSA analysis corroborates the MD simulation result and showed the lowest energy for the compounds 2734589 and 138968421. The DFT and MEP analysis also predicts the better reactivity and stability of both the hit compounds. Overall, both andrographolide derivatives exhibit good potential as potent inhibitors for both nsp14 and nsp16 proteins, however, in-vitro and in vivo assessment would be required to prove their efficacy and safety in clinical settings. Moreover, the drug discovery strategy aiming at the dual target approach might serve as a useful model for inventing novel drug molecules for various other diseases.
Author contributions J.T. and J.S. contributed to the study conceptualization and design and were involved in material preparation, data collection and analysis. A.G. and S.R. were involved in molecular dynamics simulations study for providing computational server access. The initial main draft of the manuscript was written by J.T., with additions and revisions from J.S. and S.R. All the authors read and approved the final manuscript.
Competing interests The authors declare no competing interests. Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Abraham, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX
Baell, Holloway, New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays, J. Med. Chem
Banerjee, Eckert, Schrey, Preissner, ProTox-II: A webserver for the prediction of toxicity of chemicals, Nucleic Acids Res
Becares, Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response, J. Virol
Chandra, Chaudhary, Qamar, Singh, Nain, In silico identification and validation of natural antiviral compounds as potential inhibitors of SARS-CoV-2 methyltransferase, J. Biomol. Struct. Dyn
Chen, Zhang, Li, Han, Probing ligand-binding modes and binding mechanisms of benzoxazole-based amide inhibitors with soluble epoxide hydrolase by molecular docking and molecular dynamics simulation, J. Phys. Chem. B
Czarna, Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity, Structure
Dennington, Keith, Millam, Inc, Shawnee Mission et al., None, Version
Dyall, Middle east respiratory syndrome and severe acute respiratory syndrome: Current therapeutic options and potential targets for novel therapies, Drugs
Hanna, Informatics and computational approaches for the discovery and optimization of natural product-inspired inhibitors of the SARS-CoV-2 2′-O-methyltransferase, J. Nat. Prod
Hossain, Urbi, Sule, Rahman, Andrographis Paniculata (burm et al., ex Nees: A review of ethnobotany, phytochemistry, and pharmacology, Sci. World J
Hosseini, Askari, A review of neurological side effects of Covid-19 vaccination, Eur. J. Med. Res
Huang, CHARMM36m: An improved force field for folded and intrinsically disordered proteins, Nat. Methods
Jain, Satija, Sudandiradoss, Discovery of andrographolide hit analog as a potent cyclooxygenase-2 inhibitor through consensus MD-simulation, electrostatic potential energy simulation and ligand efficiency metrics, Sci. Rep
Joshi, Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1760137
Kadioglu, Saeed, Greten, Efferth, Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning, Comput. Biol. Med
Kumar, Extraction, isolation, synthesis, and biological evaluation of novel piperic acid derivatives for the treatment of Alzheimer's disease, Mol. Divers, doi:10.1007/s11030-023-10667-x
Kumar, Identification and structural studies of natural inhibitors against SARS-CoV-2 viral RNA methyltransferase (NSP16), J. Biomol. Struct. Dyn
Laksmiani, Active compounds activity from the medicinal plants against SARS-CoV-2 using in silico assay, Biomed. Pharmacol. J
Li, Hilgenfeld, Whitley, De Clercq, Therapeutic strategies for Covid-19: Progress and lessons learned, Nat. Rev. Drug Discov
Li, Probing ligand binding modes of human cytochrome P450 2J2 by homology modeling, molecular dynamics simulation, and flexible molecular docking, Proteins Struct. Funct. Bioinform
Livingston, Bucher, Rekito, Coronavirus disease 2019 and influenza 2019-2020, JAMA
Moeller, Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN, Proc. Natl. Acad. Sci. U. S. A
Morris, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem
Murugan, Pandian, Jeyakanthan, Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials, J. Biomol. Struct. Dyn
Newman, Drug delivery to the lungs: Challenges and opportunities, Ther. Deliv
O'boyle, Open Babel: An Open chemical toolbox, J. Cheminform
Pholphana, Rangkadilok, Saehun, Ritruechai, Satayavivad, Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm.f.) Nees (Chuanxinlian), Chin. Med. (United Kingdom)
Rai, Molecular docking, binding mode analysis, molecular dynamics, and prediction of ADMET/toxicity properties of selective potential antiviral agents against SARS-CoV-2 main protease: An effort toward drug repurposing to combat Covid-19, Mol. Divers
Robson, Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans, Comput. Biol. Med
Romano, Ruggiero, Squeglia, Maga, Berisio, A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping, Cells
Rosas-Lemus, High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design, Sci. Signal
Shi, Wen, Song, Wang, Yu, Computational investigation of potent inhibitors against SARS-CoV-2 2′-O-methyltransferase (nsp16): Structure-based pharmacophore modeling, molecular docking, molecular dynamics simulations and binding free energy calculations, J. Mol. Graph. Model
Tahir, Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target, J. Med. Virol
Thomas, Kumar, Satija, Integrated molecular and quantum mechanical approach to identify novel potent natural bioactive compound against 2′-O-methyltransferase (nsp16) of SARS-CoV-2, J. Biomol. Struct. Dyn
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, J. Comput. Chem
Valdés-Tresanco, Valdés-Tresanco, Valiente, Moreno, Gmx_Mmpbsa, A new tool to perform end-state free energy calculations with GROMACS, J. Chem. Theory Comput
Viswanathan, Structural basis of RNA cap modification by SARS-CoV-2, Nat. Commun
Wu, Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharm. Sin. B
Yap, PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints, J. Comput. Chem
Zheng, SARS-coV-2: An emerging coronavirus that causes a global threat, Int. J. Biol. Sci
Zmudzinski, Ebselen derivatives inhibit SARS-CoV-2 replication by inhibition of its essential proteins: PLpro and Mpro proteases, and nsp14 guanine N7-methyltransferase, Sci. Rep
DOI record:
{
"DOI": "10.1038/s41598-024-58532-7",
"ISSN": [
"2045-2322"
],
"URL": "http://dx.doi.org/10.1038/s41598-024-58532-7",
"abstract": "<jats:title>Abstract</jats:title><jats:p>The Covid-19 pandemic outbreak has accelerated tremendous efforts to discover a therapeutic strategy that targets severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to control viral infection. Various viral proteins have been identified as potential drug targets, however, to date, no specific therapeutic cure is available against the SARS-CoV-2. To address this issue, the present work reports a systematic cheminformatic approach to identify the potent andrographolide derivatives that can target methyltransferases of SARS-CoV-2, i.e. nsp14 and nsp16 which are crucial for the replication of the virus and host immune evasion. A consensus of cheminformatics methodologies including virtual screening, molecular docking, ADMET profiling, molecular dynamics simulations, free-energy landscape analysis, molecular mechanics generalized born surface area (MM-GBSA), and density functional theory (DFT) was utilized. Our study reveals two new andrographolide derivatives (PubChem CID: 2734589 and 138968421) as natural bioactive molecules that can form stable complexes with both proteins via hydrophobic interactions, hydrogen bonds and electrostatic interactions. The toxicity analysis predicts class four toxicity for both compounds with LD<jats:sub>50</jats:sub> value in the range of 500–700 mg/kg. MD simulation reveals the stable formation of the complex for both the compounds and their average trajectory values were found to be lower than the control inhibitor and protein alone. MMGBSA analysis corroborates the MD simulation result and showed the lowest energy for the compounds 2734589 and 138968421. The DFT and MEP analysis also predicts the better reactivity and stability of both the hit compounds. Overall, both andrographolide derivatives exhibit good potential as potent inhibitors for both nsp14 and nsp16 proteins, however, in-vitro and in vivo assessment would be required to prove their efficacy and safety in clinical settings. Moreover, the drug discovery strategy aiming at the dual target approach might serve as a useful model for inventing novel drug molecules for various other diseases.</jats:p>",
"alternative-id": [
"58532"
],
"article-number": "9801",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "22 December 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "1 April 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "29 April 2024"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Thomas",
"given": "Jobin",
"sequence": "first"
},
{
"affiliation": [],
"family": "Ghosh",
"given": "Anupam",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ranjan",
"given": "Shivendu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Satija",
"given": "Jitendra",
"sequence": "additional"
}
],
"container-title": "Scientific Reports",
"container-title-short": "Sci Rep",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T11:03:01Z",
"timestamp": 1714388581000
},
"deposited": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T11:03:09Z",
"timestamp": 1714388589000
},
"indexed": {
"date-parts": [
[
2024,
4,
30
]
],
"date-time": "2024-04-30T00:26:52Z",
"timestamp": 1714436812836
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
4,
29
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T00:00:00Z",
"timestamp": 1714348800000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T00:00:00Z",
"timestamp": 1714348800000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41598-024-58532-7.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41598-024-58532-7",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41598-024-58532-7.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2024,
4,
29
]
]
},
"published-online": {
"date-parts": [
[
2024,
4,
29
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1001/jama.2020.2633",
"author": "E Livingston",
"doi-asserted-by": "publisher",
"first-page": "1122",
"journal-title": "JAMA",
"key": "58532_CR1",
"unstructured": "Livingston, E., Bucher, K. & Rekito, A. Coronavirus disease 2019 and influenza 2019–2020. JAMA 323, 1122–1122 (2020).",
"volume": "323",
"year": "2020"
},
{
"DOI": "10.1007/s40265-017-0830-1",
"author": "J Dyall",
"doi-asserted-by": "publisher",
"first-page": "1935",
"journal-title": "Drugs",
"key": "58532_CR2",
"unstructured": "Dyall, J. et al. Middle east respiratory syndrome and severe acute respiratory syndrome: Current therapeutic options and potential targets for novel therapies. Drugs 77, 1935–1966 (2017).",
"volume": "77",
"year": "2017"
},
{
"DOI": "10.7150/ijbs.45053",
"author": "J Zheng",
"doi-asserted-by": "publisher",
"first-page": "1678",
"journal-title": "Int. J. Biol. Sci.",
"key": "58532_CR3",
"unstructured": "Zheng, J. SARS-coV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 16, 1678–1685 (2020).",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1186/s40001-023-00992-0",
"author": "R Hosseini",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Eur. J. Med. Res.",
"key": "58532_CR4",
"unstructured": "Hosseini, R. & Askari, N. A review of neurological side effects of Covid-19 vaccination. Eur. J. Med. Res. 28, 1–8 (2023).",
"volume": "28",
"year": "2023"
},
{
"DOI": "10.1016/j.compbiomed.2020.103849",
"author": "B Robson",
"doi-asserted-by": "publisher",
"journal-title": "Comput. Biol. Med.",
"key": "58532_CR5",
"unstructured": "Robson, B. Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans. Comput. Biol. Med. 122, 103849 (2020).",
"volume": "122",
"year": "2020"
},
{
"DOI": "10.1080/07391102.2020.1760137",
"author": "RS Joshi",
"doi-asserted-by": "publisher",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "58532_CR6",
"unstructured": "Joshi, R. S. et al. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2020.1760137 (2020).",
"year": "2020"
},
{
"DOI": "10.1007/s11030-021-10188-5",
"author": "H Rai",
"doi-asserted-by": "publisher",
"first-page": "1905",
"journal-title": "Mol. Divers.",
"key": "58532_CR7",
"unstructured": "Rai, H. et al. Molecular docking, binding mode analysis, molecular dynamics, and prediction of ADMET/toxicity properties of selective potential antiviral agents against SARS-CoV-2 main protease: An effort toward drug repurposing to combat Covid-19. Mol. Divers. 25, 1905–1927 (2021).",
"volume": "25",
"year": "2021"
},
{
"DOI": "10.1016/j.apsb.2020.02.008",
"author": "C Wu",
"doi-asserted-by": "publisher",
"first-page": "766",
"journal-title": "Acta Pharm. Sin. B",
"key": "58532_CR8",
"unstructured": "Wu, C. et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 10, 766–788 (2020).",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"author": "G Li",
"doi-asserted-by": "publisher",
"first-page": "449",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "58532_CR9",
"unstructured": "Li, G., Hilgenfeld, R., Whitley, R. & De Clercq, E. Therapeutic strategies for Covid-19: Progress and lessons learned. Nat. Rev. Drug Discov. 22, 449–475 (2023).",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.1080/07391102.2023.2206287",
"author": "J Thomas",
"doi-asserted-by": "publisher",
"first-page": "1999",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "58532_CR10",
"unstructured": "Thomas, J., Kumar, S. & Satija, J. Integrated molecular and quantum mechanical approach to identify novel potent natural bioactive compound against 2′-O-methyltransferase (nsp16) of SARS-CoV-2. J. Biomol. Struct. Dyn. 42, 1999–2012 (2023).",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1080/07391102.2021.1997821",
"author": "M Kumar",
"doi-asserted-by": "publisher",
"first-page": "13965",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "58532_CR11",
"unstructured": "Kumar, M. et al. Identification and structural studies of natural inhibitors against SARS-CoV-2 viral RNA methyltransferase (NSP16). J. Biomol. Struct. Dyn. 40, 13965–13975 (2021).",
"volume": "40",
"year": "2021"
},
{
"DOI": "10.1002/jmv.27009",
"author": "M Tahir",
"doi-asserted-by": "publisher",
"first-page": "4258",
"journal-title": "J. Med. Virol.",
"key": "58532_CR12",
"unstructured": "Tahir, M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J. Med. Virol. 93, 4258–4264 (2021).",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1073/pnas.2106379119",
"author": "NH Moeller",
"doi-asserted-by": "publisher",
"journal-title": "Proc. Natl. Acad. Sci. U. S. A.",
"key": "58532_CR13",
"unstructured": "Moeller, N. H. et al. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. U. S. A. 119, e2106379119 (2022).",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1038/s41467-020-17496-8",
"author": "T Viswanathan",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Nat. Commun.",
"key": "58532_CR14",
"unstructured": "Viswanathan, T. et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 11, 1–7 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41598-023-35907-w",
"author": "M Zmudzinski",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Sci. Rep.",
"key": "58532_CR15",
"unstructured": "Zmudzinski, M. et al. Ebselen derivatives inhibit SARS-CoV-2 replication by inhibition of its essential proteins: PLpro and Mpro proteases, and nsp14 guanine N7-methyltransferase. Sci. Rep. 13, 1–16 (2023).",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1016/j.jmgm.2022.108306",
"author": "L Shi",
"doi-asserted-by": "publisher",
"journal-title": "J. Mol. Graph. Model.",
"key": "58532_CR16",
"unstructured": "Shi, L., Wen, Z., Song, Y., Wang, J. & Yu, D. Computational investigation of potent inhibitors against SARS-CoV-2 2′-O-methyltransferase (nsp16): Structure-based pharmacophore modeling, molecular docking, molecular dynamics simulations and binding free energy calculations. J. Mol. Graph. Model. 117, 108306 (2022).",
"volume": "117",
"year": "2022"
},
{
"DOI": "10.1021/acs.jnatprod.3c00875",
"author": "GS Hanna",
"doi-asserted-by": "publisher",
"first-page": "217",
"journal-title": "J. Nat. Prod.",
"key": "58532_CR17",
"unstructured": "Hanna, G. S. et al. Informatics and computational approaches for the discovery and optimization of natural product-inspired inhibitors of the SARS-CoV-2 2′-O-methyltransferase. J. Nat. Prod. 87, 217–227 (2023).",
"volume": "87",
"year": "2023"
},
{
"DOI": "10.1080/07391102.2021.1886174",
"author": "A Chandra",
"doi-asserted-by": "publisher",
"first-page": "6534",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "58532_CR18",
"unstructured": "Chandra, A., Chaudhary, M., Qamar, I., Singh, N. & Nain, V. In silico identification and validation of natural antiviral compounds as potential inhibitors of SARS-CoV-2 methyltransferase. J. Biomol. Struct. Dyn. 40, 6534–6544 (2022).",
"volume": "40",
"year": "2022"
},
{
"author": "N Pholphana",
"first-page": "1",
"journal-title": "Chin. Med. (United Kingdom)",
"key": "58532_CR19",
"unstructured": "Pholphana, N., Rangkadilok, N., Saehun, J., Ritruechai, S. & Satayavivad, J. Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm.f.) Nees (Chuanxinlian). Chin. Med. (United Kingdom) 8, 1–12 (2013).",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.1155/2014/274905",
"author": "MS Hossain",
"doi-asserted-by": "publisher",
"first-page": "274905",
"journal-title": "Sci. World J.",
"key": "58532_CR20",
"unstructured": "Hossain, M. S., Urbi, Z., Sule, A. & Rahman, K. M. H. Andrographis paniculata (Burm. f.) Wall. ex Nees: A review of ethnobotany, phytochemistry, and pharmacology. Sci. World J. 2014, 274905 (2014).",
"volume": "2014",
"year": "2014"
},
{
"DOI": "10.1080/07391102.2020.1777901",
"author": "NA Murugan",
"doi-asserted-by": "publisher",
"first-page": "4415",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "58532_CR21",
"unstructured": "Murugan, N. A., Pandian, C. J. & Jeyakanthan, J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J. Biomol. Struct. Dyn. 39, 4415–4426 (2020).",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1038/s41598-023-35192-7",
"author": "P Jain",
"doi-asserted-by": "publisher",
"first-page": "8147",
"journal-title": "Sci. Rep.",
"key": "58532_CR22",
"unstructured": "Jain, P., Satija, J. & Sudandiradoss, C. Discovery of andrographolide hit analog as a potent cyclooxygenase-2 inhibitor through consensus MD-simulation, electrostatic potential energy simulation and ligand efficiency metrics. Sci. Rep. 13, 8147 (2023).",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.13005/bpj/1953",
"author": "NP Linda Laksmiani",
"doi-asserted-by": "publisher",
"first-page": "873",
"journal-title": "Biomed. Pharmacol. J.",
"key": "58532_CR23",
"unstructured": "Linda Laksmiani, N. P. et al. Active compounds activity from the medicinal plants against SARS-CoV-2 using in silico assay. Biomed. Pharmacol. J. 13, 873–881 (2020).",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1016/j.compbiomed.2021.104359",
"author": "O Kadioglu",
"doi-asserted-by": "publisher",
"journal-title": "Comput. Biol. Med.",
"key": "58532_CR24",
"unstructured": "Kadioglu, O., Saeed, M., Greten, H. J. & Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Comput. Biol. Med. 133, 104359 (2021).",
"volume": "133",
"year": "2021"
},
{
"DOI": "10.1016/j.str.2022.04.014",
"author": "A Czarna",
"doi-asserted-by": "publisher",
"first-page": "1050",
"journal-title": "Structure",
"key": "58532_CR25",
"unstructured": "Czarna, A. et al. Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity. Structure 30, 1050–1054 (2022).",
"volume": "30",
"year": "2022"
},
{
"DOI": "10.1126/scisignal.abe1202",
"author": "M Rosas-Lemus",
"doi-asserted-by": "publisher",
"first-page": "1202",
"journal-title": "Sci. Signal.",
"key": "58532_CR26",
"unstructured": "Rosas-Lemus, M. et al. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 13, 1202 (2020).",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1186/1758-2946-3-1",
"author": "NM O’Boyle",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "J. Cheminform.",
"key": "58532_CR27",
"unstructured": "O’Boyle, N. M. et al. Open Babel: An Open chemical toolbox. J. Cheminform. 3, 1–14 (2011).",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.1002/jcc.21334",
"author": "O Trott",
"doi-asserted-by": "publisher",
"first-page": "455",
"journal-title": "J. Comput. Chem.",
"key": "58532_CR28",
"unstructured": "Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1128/JVI.03259-15",
"author": "M Becares",
"doi-asserted-by": "publisher",
"first-page": "5399",
"journal-title": "J. Virol.",
"key": "58532_CR29",
"unstructured": "Becares, M. et al. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J. Virol. 90, 5399–5414 (2016).",
"volume": "90",
"year": "2016"
},
{
"DOI": "10.3390/cells9051267",
"author": "M Romano",
"doi-asserted-by": "publisher",
"first-page": "1267",
"journal-title": "Cells",
"key": "58532_CR30",
"unstructured": "Romano, M., Ruggiero, A., Squeglia, F., Maga, G. & Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 9, 1267 (2020).",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1002/jcc.21256",
"author": "GM Morris",
"doi-asserted-by": "publisher",
"first-page": "2785",
"journal-title": "J. Comput. Chem.",
"key": "58532_CR31",
"unstructured": "Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).",
"volume": "30",
"year": "2009"
},
{
"DOI": "10.1002/jcc.21707",
"author": "CW Yap",
"doi-asserted-by": "publisher",
"first-page": "1466",
"journal-title": "J. Comput. Chem.",
"key": "58532_CR32",
"unstructured": "Yap, C. W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474 (2011).",
"volume": "32",
"year": "2011"
},
{
"DOI": "10.1093/nar/gky318",
"author": "P Banerjee",
"doi-asserted-by": "publisher",
"first-page": "W257",
"journal-title": "Nucleic Acids Res.",
"key": "58532_CR33",
"unstructured": "Banerjee, P., Eckert, A. O., Schrey, A. K. & Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263 (2018).",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.1021/jm901137j",
"author": "JB Baell",
"doi-asserted-by": "publisher",
"first-page": "2719",
"journal-title": "J. Med. Chem.",
"key": "58532_CR34",
"unstructured": "Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).",
"volume": "53",
"year": "2010"
},
{
"DOI": "10.1016/j.softx.2015.06.001",
"author": "MJ Abraham",
"doi-asserted-by": "publisher",
"first-page": "19",
"journal-title": "SoftwareX",
"key": "58532_CR35",
"unstructured": "Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).",
"volume": "1–2",
"year": "2015"
},
{
"DOI": "10.1038/nmeth.4067",
"author": "J Huang",
"doi-asserted-by": "publisher",
"first-page": "71",
"journal-title": "Nat. Methods",
"key": "58532_CR36",
"unstructured": "Huang, J. et al. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2016).",
"volume": "14",
"year": "2016"
},
{
"DOI": "10.1021/acs.jctc.1c00645",
"author": "MS Valdés-Tresanco",
"doi-asserted-by": "publisher",
"first-page": "6281",
"journal-title": "J. Chem. Theory Comput.",
"key": "58532_CR37",
"unstructured": "Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A. & Moreno, E. Gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291 (2021).",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1007/s11030-023-10667-x",
"author": "J Kumar",
"doi-asserted-by": "publisher",
"journal-title": "Mol. Divers.",
"key": "58532_CR38",
"unstructured": "Kumar, J. et al. Extraction, isolation, synthesis, and biological evaluation of novel piperic acid derivatives for the treatment of Alzheimer’s disease. Mol. Divers. https://doi.org/10.1007/s11030-023-10667-x (2023).",
"year": "2023"
},
{
"author": "R Dennington",
"first-page": "1",
"journal-title": "Version",
"key": "58532_CR39",
"unstructured": "Dennington, R., Keith, T. A., Millam, J. M., Inc, S. & Shawnee Mission, K. S. GaussView. Version 6, 1 (2016).",
"volume": "6",
"year": "2016"
},
{
"DOI": "10.4155/tde-2017-0037",
"author": "SP Newman",
"doi-asserted-by": "publisher",
"first-page": "647",
"journal-title": "Ther. Deliv.",
"key": "58532_CR40",
"unstructured": "Newman, S. P. Drug delivery to the lungs: Challenges and opportunities. Ther. Deliv. 8, 647–661 (2017).",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.1021/jp304736e",
"author": "H Chen",
"doi-asserted-by": "publisher",
"first-page": "10219",
"journal-title": "J. Phys. Chem. B",
"key": "58532_CR41",
"unstructured": "Chen, H., Zhang, Y., Li, L. & Han, J. G. Probing ligand-binding modes and binding mechanisms of benzoxazole-based amide inhibitors with soluble epoxide hydrolase by molecular docking and molecular dynamics simulation. J. Phys. Chem. B 116, 10219–10233 (2012).",
"volume": "116",
"year": "2012"
},
{
"DOI": "10.1002/prot.21778",
"author": "W Li",
"doi-asserted-by": "publisher",
"first-page": "938",
"journal-title": "Proteins Struct. Funct. Bioinform.",
"key": "58532_CR42",
"unstructured": "Li, W. et al. Probing ligand binding modes of human cytochrome P450 2J2 by homology modeling, molecular dynamics simulation, and flexible molecular docking. Proteins Struct. Funct. Bioinform. 71, 938–949 (2008).",
"volume": "71",
"year": "2008"
}
],
"reference-count": 42,
"references-count": 42,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41598-024-58532-7"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "14"
}