Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus
et al., International Journal of Molecular Sciences, doi:10.3390/ijms231911131, Sep 2022
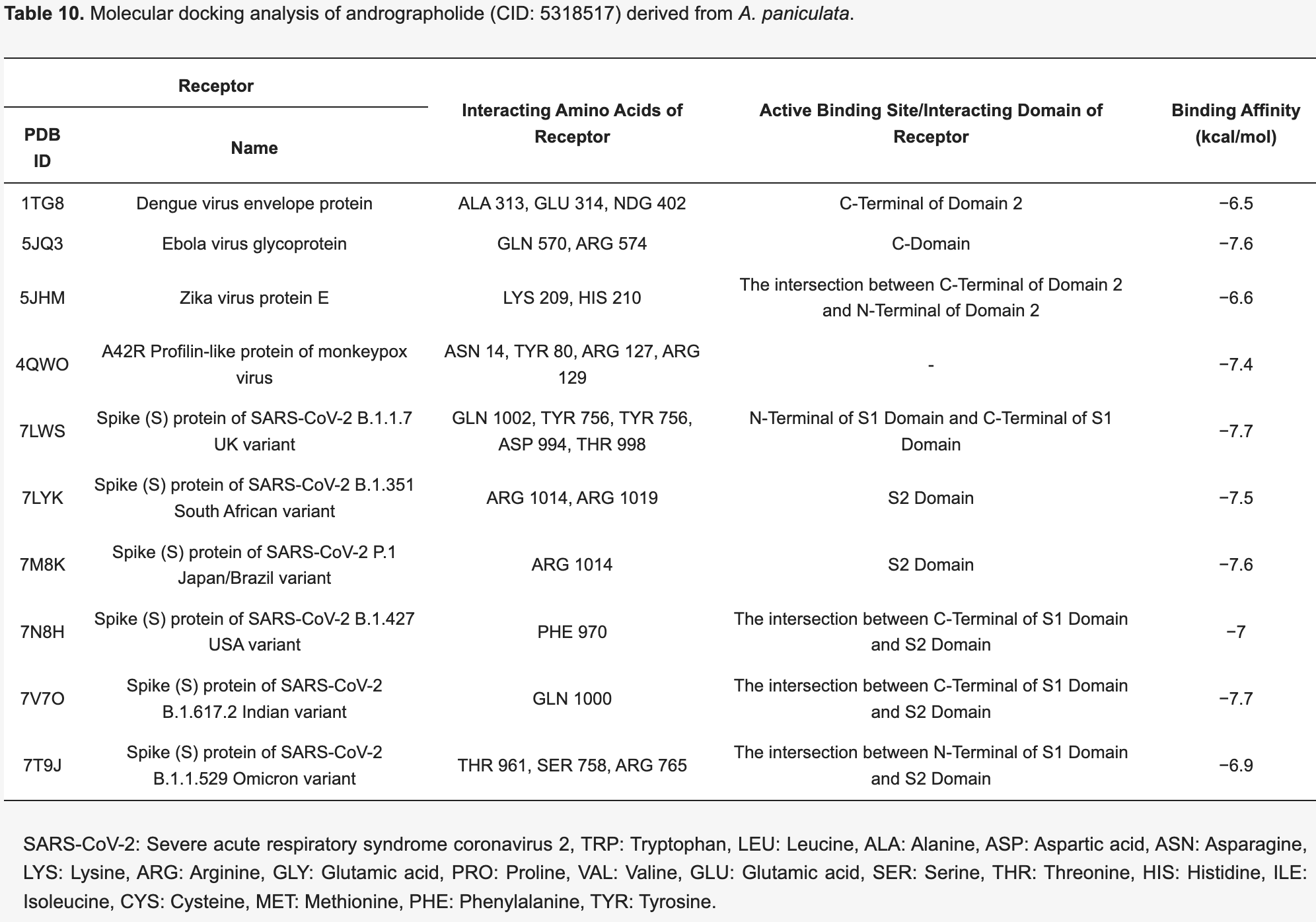
In silico study showing that the phytochemical andrographolide derived from Andrographis paniculata binds strongly to the spike proteins of six SARS-CoV-2 variants, including the B.1.1.7 UK, B.1.351 South African, P.1 Japan/Brazil, B.1.427 USA, B.1.617.2 Indian, and B.1.1.529 Omicron variants, with binding energies ranging from -6.9 to -7.7 kcal/mol.
25 preclinical studies support the efficacy of andrographolide for COVID-19:
In vitro studies demonstrate inhibition of the MproA,18 protein.
In vitro studies demonstrate efficacy in Calu-3B,18, A549C,14, and HUVECD,18 cells.
Animal studies demonstrate efficacy in Sprague Dawley miceE,18 and Golden Syrian hamstersF,14.
Andrographolide inhibits Mpro in a dose-dependent manner18, reduces ACE2 levels in the lung tissue of mice in combination with baicalein18, inhibits binding between the SARS-CoV-2 spike protein and ACE218, alleviates lung inflammation and cytokine storm in mice18, and improves survival and reduces lung inflammation via anti-inflammatory effects in Syrian hamsters14.
1.
Zhang et al., Effects and Mechanisms of Andrographolide for COVID-19: A Network Pharmacology-Based and Experimentally Validated Study, Natural Product Communications, doi:10.1177/1934578X241288428.
2.
Thomas et al., Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-58532-7.
3.
Arifin et al., Computational exploration of Andrographis paniculata herb compounds as potential antiviral agents targeting NSP3 (6W02) and NSP5 (7AR6) of SARS-COV-2, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2023.25.2.0292.
4.
Bhattarai et al., Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2174596.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
6.
Dassanayake et al., Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus, International Journal of Molecular Sciences, doi:10.3390/ijms231911131.
7.
Ningrum et al., Potency Of Andrographolide, L-Mimosine And Asiaticoside Compound As Antiviral For Covid-19 Based On In Silico Method, Proceedings Universitas Muhammadiyah Yogyakarta Undergraduate Conference, doi:10.18196/umygrace.v2i2.418.
8.
Ravichandran et al., Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach, Biointerface Research in Applied Chemistry, doi:10.33263/BRIAC132.155.
9.
Saeheng et al., In Silico Prediction of Andrographolide Dosage Regimens for COVID-19 Treatment, The American Journal of Chinese Medicine, doi:10.1142/S0192415X22500732.
10.
Khanal et al., Combination of system biology to probe the anti-viral activity of andrographolide and its derivative against COVID-19, RSC Advances, doi:10.1039/D0RA10529E.
11.
Rehan et al., A Computational Approach Identified Andrographolide as a Potential Drug for Suppressing COVID-19-Induced Cytokine Storm, Frontiers in Immunology, doi:10.3389/fimmu.2021.648250.
12.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
13.
Dey et al., The role of andrographolide and its derivative in COVID-19 associated proteins and immune system, Research Square, doi:10.21203/rs.3.rs-35800/v1.
14.
Kongsomros et al., In vivo evaluation of Andrographis paniculata and Boesenbergia rotunda extract activity against SARS-CoV-2 Delta variant in Golden Syrian hamsters: Potential herbal alternative for COVID-19 treatment, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2024.05.004.
15.
Chaopreecha et al., Andrographolide attenuates SARS-CoV-2 infection via an up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC), Phytomedicine, doi:10.1016/j.phymed.2024.156279.
16.
Li et al., Andrographolide suppresses SARS-CoV-2 infection by downregulating ACE2 expression: A mechanistic study, Antiviral Therapy, doi:10.1177/13596535241259952.
17.
Low et al., The wide spectrum anti-inflammatory activity of andrographolide in comparison to NSAIDs: a promising therapeutic compound against the cytokine storm, bioRxiv, doi:10.1101/2024.02.21.581396.
18.
Wan et al., Synergistic inhibition effects of andrographolide and baicalin on coronavirus mechanisms by downregulation of ACE2 protein level, Scientific Reports, doi:10.1038/s41598-024-54722-5.
19.
Siridechakorn et al., Inhibitory efficiency of Andrographis paniculata extract on viral multiplication and nitric oxide production, Scientific Reports, doi:10.1038/s41598-023-46249-y.
a.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
b.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
c.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
d.
HUVEC (Human Umbilical Vein Endothelial Cells) are primary endothelial cells derived from the vein of the umbilical cord. They are used to study vascular biology, including inflammation, angiogenesis, and viral interactions with endothelial cells.
e.
An outbred multipurpose breed of albino mouse used extensively in medical research.
f.
A rodent model widely used in infectious disease research due to their susceptibility to viral infections and similar disease progression to humans.
Dassanayake et al., 22 Sep 2022, peer-reviewed, 4 authors.
Contact: tengjin.khoo@nottingham.edu.my (corresponding author).
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus
International Journal of Molecular Sciences, doi:10.3390/ijms231911131
The emergence and rapid evolution of human pathogenic viruses, combined with the difficulties in developing effective vaccines, underline the need to develop innovative broad-spectrum antiviral therapeutic agents. The present study aims to determine the in silico antiviral potential of six bacterial antimicrobial peptides (AMPs), two phytochemicals (silvestrol, andrographolide), and two bacterial secondary metabolites (lyngbyabellin A, hapalindole H) against dengue virus, Zika virus, Ebola virus, the major variants of SARS-CoV-2 and monkeypox virus. The comparison of docking scores obtained with natural biomolecules was performed with specific neutralizing antibodies (positive controls for ClusPro) and antiviral drugs (negative controls for Autodock Vina). Glycocin F was the only natural biomolecule tested to show high binding energies to all viral surface proteins and the corresponding viral cell receptors. Lactococcin G and plantaricin ASM1 also achieved high docking scores with all viral surface proteins and most corresponding cell surface receptors. Silvestrol, andrographolide, hapalindole H, and lyngbyabellin A showed variable docking scores depending on the viral surface proteins and cell receptors tested. Three glycocin F mutants with amino acid modifications showed an increase in their docking energy to the spike proteins of SARS-CoV-2 B.1.617.2 Indian variant, and of the SARS-CoV-2 P.1 Japan/Brazil variant, and the dengue DENV envelope protein. All mutant AMPs indicated a frequent occurrence of valine and proline amino acid rotamers. AMPs and glycocin F in particular are the most promising biomolecules for the development of broad-spectrum antiviral treatments targeting the attachment and entry of viruses into their target cell.
References
Adalja, Inglesby, Broad-Spectrum Antiviral Agents: A Crucial Pandemic Tool, Expert Rev. Anti-Infect. Ther, doi:10.1080/14787210.2019.1635009
Adiguna, Panggabean, Atikana, Untari, Izzati et al., Antiviral and Immunostimulant Activities of Andrographis paniculata, HAYATI J. Biosci, doi:10.4308/hjb.22.2.67
Al-Tawfiq, Barry, Memish, International outbreaks of Monkeypox virus infection with no established travel: A public health concern with significant knowledge gap, Travel Med. Infect. Dis, doi:10.1016/j.tmaid.2022.102364
Aminu, Ibrahim, Sallau, Interaction of SARS-CoV-2 spike protein with angiotensin converting enzyme inhibitors and selected compounds from the chemical entities of biological interest, Beni-Suef Univ. J. Basic Appl. Sci, doi:10.1186/s43088-021-00138-3
Ansah, Matchar, Shao, Low, Pourghaderi et al., The effectiveness of public health interventions against COVID-19: Lessons from the Singapore experience, PLoS ONE, doi:10.1371/journal.pone.0248742
Artese, Svicher, Costa, Salpini, Di Maio et al., Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses, Drug Resist, doi:10.1016/j.drup.2020.100721
Balmeh, Mahmoundi, Fard, Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease, Inform. Med. Unlocked, doi:10.1016/j.imu.2021.100515
Bansal, Mohagaonkar, Sen, Khanam, Rathi, In-silico study of peptide-protein interaction of antimicrobial peptides potentially targeting SARS and SARS-CoV-2 nucleocapsid protein, Silico Pharmacol, doi:10.1007/s40203-021-00103-z
Bickley, Chan, Skali, Stadelmann, Torgler, How does globalization affect COVID-19 responses?, Glob. Health, doi:10.1186/s12992-021-00677-5
Biedenkopf, Lange-Grünweller, Schulte, Weißer, Müller et al., The natural compound silvestrol is a potent inhibitor of Ebola virus replication, Antivir. Res, doi:10.1016/j.antiviral.2016.11.011
Burrell, Epidemiology of Viral Infections, doi:10.1016/B978-0-12-375156-0.00013-8
Chen, Hoover, Bacteriocins and their Food Applications, Compr. Rev. Food Sci. Food Saf, doi:10.1111/j.1541-4337.2003.tb00016.x
Comeau, Gatchell, Vajda, Camacho, Cluspro, A fully automated algorithm for protein-protein docking, Nucleic Acids Res, doi:10.1093/nar/gkh354
Dai, Gao, Viral targets for vaccines against COVID-19, Nat. Rev. Immunol, doi:10.1038/s41577-020-00480-0
Desta, Porter, Xia, Kozakov, Vajda, Performance and Its Limits in Rigid Body Protein-Protein Docking, Structure, doi:10.1016/j.str.2020.06.006
Dhama, Khan, Tiwari, Sircar, Bhat et al., Coronavirus Disease 2019-COVID-19, Clin. Microbiol. Rev, doi:10.1128/CMR.00028-20
El-Baz, El-Senousy, El-Sayed, Kamel, In vitro antiviral and antimicrobial activities of Spirulina platensis extract, J. Appl. Pharm. Sci, doi:10.7324/JAPS.2013.31209
Elgner, Sabino, Basic, Ploen, Grünweller et al., Inhibition of Zika Virus Replication by Silvestrol, Viruses, doi:10.3390/v10040149
Elongngono, Shresta, Immune Response to Dengue and Zika, Annu. Rev. Immunol, doi:10.1146/annurev-immunol-042617-053142
Enmozhi, Raja, Sebastine, Joseph, Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2020.1760136
Gagnière, Di Martino, Protein and peptide research applied to Covid-19 and SARS-CoV-2, Open Access Res. J. Biol. Pharm, doi:10.53022/oarjbp.2021.2.1.0037
Geraghty, Aliota, Bonnac, Broad-Spectrum Antiviral Strategies and Nucleoside Analogues, Viruses, doi:10.3390/v13040667
Ghildiyal, Prakash, Chaudhary, Gupta, Gabrani, Phytochemicals as Antiviral Agents: Recent Updates, doi:10.1007/978-981-15-1761-7_12
Gil, Ginex, Maestro, Nozal, Barrado-Gil et al., COVID-19: Drug Targets and Potential Treatments, J. Med. Chem, doi:10.1021/acs.jmedchem.0c00606
Grubaugh, Ladner, Lemey, Pybus, Rambaut et al., Tracking virus outbreaks in the twenty-first century, Nat. Microbiol, doi:10.1038/s41564-018-0296-2
Harwansh, Bahadur, Herbal Medicines to Fight Against COVID-19: New Battle with an Old Weapon, Curr. Pharm. Biotechnol, doi:10.2174/1389201022666210322124348
Henss, Scholz, Grünweller, Schnierle, Silvestrol Inhibits Chikungunya Virus Replication, Viruses, doi:10.3390/v10110592
Hickman, Saunders, Bigger, Kane, Iversen, The development of broad-spectrum antiviral medical countermeasures to treat viral hemorrhagic fevers caused by natural or weaponized virus infections, PLoS Negl. Trop. Dis, doi:10.1371/journal.pntd.0010220
Hiremath, Kumar, Nandan, Mantesh, Shankarappa et al., In silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in Ayurveda medicine in inhibiting SARS-CoV-2, Biotech, doi:10.1007/s13205-020-02578-7
Hoffmann, Guha, Wu, Ghimire, Wang et al., Broad-Spectrum Antiviral Entry Inhibition by Interfacially Active Peptides, J. Virol, doi:10.1128/JVI.01682-20
Kozakov, Beglov, Bohnuud, Mottarella, Xia et al., How good is automated protein docking?, Proteins Struct. Funct. Bioinform, doi:10.1002/prot.24403
Kozakov, Hall, Xia, Porter, Padhorny et al., The ClusPro web server for proteinprotein docking, Nat. Protoc, doi:10.1038/nprot.2016.169
Li, Khanom, Sun, Paemanee, Roytrakul et al., Andrographolide and Its 14-Aryloxy Analogues Inhibit Zika and Dengue Virus Infection, Molecules, doi:10.3390/molecules25215037
Lim, Chan, Tan, Teh, Mohd et al., Ex Nees, Andrographolide, and Andrographolide Analogues as SARS-CoV-2 Antivirals? A Rapid Review, Nat. Prod. Commun, doi:10.1177/1934578X211016610
Longet, Mellors, Carroll, Tipton, Ebolavirus, Comparison of Survivor Immunology and Animal Models in the Search for a Correlate of Protection, Front. Immunol, doi:10.3389/fimmu.2020.599568
Małaczewska, Kaczorek-Łukowska, Wójcik, Siwicki, Antiviral effects of nisin, lysozyme, lactoferrin and their mixtures against bovine viral diarrhoea virus, BMC Vet Res, doi:10.1186/s12917-019-2067-6
Mirashrafi, Moravejolahkami, Balouch, Hojjati, Bahreini-Esfahani et al., The efficacy of probiotics on virus titres and antibody production in virus diseases: A systematic review on recent evidence for COVID-19 treatment, Clin. Nutr. ESPEN, doi:10.1016/j.clnesp.2021.10.016
Mulder, Lima, Miranda, Dias, Franco, Current scenario of peptide-based drugs: The key roles of cationic antitumor and antiviral peptides, Front. Microbiol, doi:10.3389/fmicb.2013.00321
Murad, Atta-Ur-Rahman, Infectious, Diseases, None
Musarra-Pizzo, Pennisi, Ben-Amor, Mandalari, Sciortino, Antiviral Activity Exerted by Natural Products against Human Viruses, Viruses, doi:10.3390/v13050828
Müller, Obermann, Karl, Wendel, Taroncher-Oldenburg et al., The rocaglate CR-31-B (-) inhibits SARS-CoV-2 replication at non-cytotoxic, low nanomolar concentrations in vitro and ex vivo, Antivir. Res, doi:10.1016/j.antiviral.2021.105012
Müller, Schulte, Lange-Grünweller, Obermann, Madhugiri et al., Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses, Antivir. Res, doi:10.1016/j.antiviral.2017.12.010
Pagarete, Ramos, Puntervoll, Allen, Verdelho, Antiviral Potential of Algal Metabolites-A Comprehensive Review, Mar. Drugs, doi:10.3390/md19020094
Panraksa, Ramphan, Khongwichit, Smith, Activity of andrographolide against dengue virus, Antivir. Res, doi:10.1016/j.antiviral.2016.12.014
Piret, Boivin, Pandemics Throughout History, Front. Microbiol, doi:10.3389/fmicb.2020.631736
Reperant, Osterhaus, Aids, Sars, Mers et al., what next?, Vaccine, doi:10.1016/j.vaccine.2017.04.082
Rohr, Barrett, Civitello, Craft, Delius et al., Emerging human infectious diseases and the links to global food production, Nat. Sustain, doi:10.1038/s41893-019-0293-3
Sa-Ngiamsuntorn, Suksatu, Pewkliang, Thongsri, Kanjanasirirat et al., Anti-SARS-CoV-2 Activity of Andrographis paniculata Extract and Its Major Component Andrographolide in Human Lung Epithelial Cells and Cytotoxicity Evaluation in Major Organ Cell Representatives, J. Nat. Prod, doi:10.1021/acs.jnatprod.0c01324
Saunders-Hastings, Krewski, Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission, Pathogens, doi:10.3390/pathogens5040066
Schrödinger, De Lano, Incentive PyMOL Software Package
Seubsasana, Pientong, Ekalaksananan, Thongchai, Aromdee, A Potential Andrographolide Analogue against the Replication of Herpes Simplex Virus Type 1 in Vero Cells, Med. Chem, doi:10.2174/157340611795564268
Shao, Li, Goraya, Wang, Chen et al., Evolution of Influenza A Virus by Mutation and Re-Assortment, Int. J. Mol. Sci, doi:10.3390/ijms18081650
Sheahan, Sims, Graham, Menachery, Gralinski et al., Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses, Sci. Transl. Med, doi:10.1126/scitranslmed.aal3653
Shityakov, Sohajda, Puskás, Roewer, Förster et al., Ionization states, cellular toxicity and molecular modeling studies of midazolam complexed with trimethyl-β-cyclodextrin, Molecules, doi:10.3390/molecules191016861
Singh, Singh, Kumar, Kabir, Kamal et al., Multi-Omics Approach in the Identification of Potential Therapeutic Biomolecule for COVID-19, Front. Pharmacol, doi:10.3389/fphar.2021.652335
Singh, Tiwari, Rai, Mohapatra, Cyanobacteria: An emerging source for drug discovery, J. Antibiot, doi:10.1038/ja.2011.21
Sliwoski, Kothiwale, Meiler, Lowe, Computational methods in drug discovery, Pharmacol. Rev, doi:10.1124/pr.112.007336
Tang, Zhu, Chen, Jiang, New technologies in computer-aided drug design: Toward target identification and new chemical entity discovery, Drug Discov. Today Technol, doi:10.1016/j.ddtec.2006.09.004
Todt, Moeller, Praditya, Kinast, Friesland et al., The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo, Antivir. Res, doi:10.1016/j.antiviral.2018.07.010
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem, doi:10.1002/jcc.21334
Vajda, Yueh, Beglov, Bohnuud, Mottarella et al., New additions to the ClusPro server motivated by CAPRI, Proteins Struct. Funct. Bioinform, doi:10.1002/prot.25219
Vallianou, Tsilingiris, Christodoulatos, Karampela, Dalamaga, Anti-viral treatment for SARS-CoV-2 infection: A race against time amidst the ongoing pandemic, Metab. Open, doi:10.1016/j.metop.2021.100096
Van De Sandt, Li, Goraya, Wang, Chen, Invasion of Influenza A Viruses from Innate and Adaptive Immune Responses, Viruses, doi:10.3390/v4091438
Wang, Wang, Lu, Qiu, Song et al., The efficacy of probiotics in patients with severe COVID-19, Ann. Palliat. Med, doi:10.21037/apm-21-3373
Wilder-Smith, COVID-19 in comparison with other emerging viral diseases: Risk of geographic spread via travel, Trop. Dis. Travel Med. Vaccines, doi:10.1186/s40794-020-00129-9
Wintachai, Kaur, Lee, Ramphan, Kuadkitkan et al., Activity of andrographolide against chikungunya virus infection, Sci. Rep, doi:10.1038/srep14179
Yuan, Zhang, Wang, Li, Wang et al., Surfactin Inhibits Membrane Fusion during Invasion of Epithelial Cells by Enveloped Viruses, J. Virol, doi:10.1128/JVI.00809-18
Zarbafian, Moghadasi, Roshandelpoor, Nan, Li et al., Protein docking refinement by convex underestimation in the low-dimensional subspace of encounter complexes, Sci. Rep, doi:10.1038/s41598-018-23982-3
Zelikin, Stellacci, Broad-Spectrum Antiviral Agents Based on Multivalent Inhibitors of Viral Infectivity, Adv. Healthc. Mater, doi:10.1002/adhm.202001433
Zhu, Meng, Wang, Wang, Broad-spectrum antiviral agents, Front. Microbiol, doi:10.3389/fmicb.2015.00517
DOI record:
{
"DOI": "10.3390/ijms231911131",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms231911131",
"abstract": "<jats:p>The emergence and rapid evolution of human pathogenic viruses, combined with the difficulties in developing effective vaccines, underline the need to develop innovative broad-spectrum antiviral therapeutic agents. The present study aims to determine the in silico antiviral potential of six bacterial antimicrobial peptides (AMPs), two phytochemicals (silvestrol, andrographolide), and two bacterial secondary metabolites (lyngbyabellin A, hapalindole H) against dengue virus, Zika virus, Ebola virus, the major variants of SARS-CoV-2 and monkeypox virus. The comparison of docking scores obtained with natural biomolecules was performed with specific neutralizing antibodies (positive controls for ClusPro) and antiviral drugs (negative controls for Autodock Vina). Glycocin F was the only natural biomolecule tested to show high binding energies to all viral surface proteins and the corresponding viral cell receptors. Lactococcin G and plantaricin ASM1 also achieved high docking scores with all viral surface proteins and most corresponding cell surface receptors. Silvestrol, andrographolide, hapalindole H, and lyngbyabellin A showed variable docking scores depending on the viral surface proteins and cell receptors tested. Three glycocin F mutants with amino acid modifications showed an increase in their docking energy to the spike proteins of SARS-CoV-2 B.1.617.2 Indian variant, and of the SARS-CoV-2 P.1 Japan/Brazil variant, and the dengue DENV envelope protein. All mutant AMPs indicated a frequent occurrence of valine and proline amino acid rotamers. AMPs and glycocin F in particular are the most promising biomolecules for the development of broad-spectrum antiviral treatments targeting the attachment and entry of viruses into their target cell.</jats:p>",
"alternative-id": [
"ijms231911131"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-3068-4349",
"affiliation": [],
"authenticated-orcid": false,
"family": "Dassanayake",
"given": "Mackingsley Kushan",
"sequence": "first"
},
{
"affiliation": [],
"family": "Khoo",
"given": "Teng-Jin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0877-1515",
"affiliation": [],
"authenticated-orcid": false,
"family": "Chong",
"given": "Chien Hwa",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5450-2733",
"affiliation": [],
"authenticated-orcid": false,
"family": "Di Martino",
"given": "Patrick",
"sequence": "additional"
}
],
"container-title": "International Journal of Molecular Sciences",
"container-title-short": "IJMS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
9,
22
]
],
"date-time": "2022-09-22T09:33:14Z",
"timestamp": 1663839194000
},
"deposited": {
"date-parts": [
[
2022,
9,
26
]
],
"date-time": "2022-09-26T04:15:57Z",
"timestamp": 1664165757000
},
"indexed": {
"date-parts": [
[
2023,
12,
27
]
],
"date-time": "2023-12-27T22:36:14Z",
"timestamp": 1703716574084
},
"is-referenced-by-count": 5,
"issue": "19",
"issued": {
"date-parts": [
[
2022,
9,
22
]
]
},
"journal-issue": {
"issue": "19",
"published-online": {
"date-parts": [
[
2022,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2022,
9,
22
]
],
"date-time": "2022-09-22T00:00:00Z",
"timestamp": 1663804800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/23/19/11131/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "11131",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2022,
9,
22
]
]
},
"published-online": {
"date-parts": [
[
2022,
9,
22
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/s41893-019-0293-3",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"DOI": "10.3389/fmicb.2020.631736",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.1038/s41564-018-0296-2",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"DOI": "10.1016/j.vaccine.2017.04.082",
"doi-asserted-by": "publisher",
"key": "ref4"
},
{
"DOI": "10.1016/j.tmaid.2022.102364",
"doi-asserted-by": "publisher",
"key": "ref5"
},
{
"DOI": "10.3390/ijms18081650",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.3390/v4091438",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1186/s40794-020-00129-9",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1016/B978-0-12-375156-0.00013-8",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1371/journal.pone.0248742",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1186/s12992-021-00677-5",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1080/14787210.2019.1635009",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.1128/JVI.01682-20",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1002/adhm.202001433",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.3390/v13040667",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.1016/j.drup.2020.100721",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.3389/fmicb.2013.00321",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1007/978-981-15-1761-7_12",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.1016/j.imu.2021.100515",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"author": "Murad",
"key": "ref20",
"series-title": "Infectious Diseases",
"year": "2021"
},
{
"DOI": "10.3390/pathogens5040066",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.1038/s41577-020-00480-0",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"DOI": "10.53022/oarjbp.2021.2.1.0037",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"DOI": "10.1002/prot.24403",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"key": "ref25",
"unstructured": "Incentive PyMOL Software Package\nhttps://pymol.org/2/"
},
{
"DOI": "10.1007/s13205-020-02578-7",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.3390/molecules191016861",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.1038/nprot.2016.169",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1016/j.metop.2021.100096",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"DOI": "10.3389/fimmu.2020.599568",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.1371/journal.pntd.0010220",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.1126/scitranslmed.aal3653",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1128/CMR.00028-20",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.3389/fmicb.2015.00517",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.1021/acs.jmedchem.0c00606",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.1146/annurev-immunol-042617-053142",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.3390/v13050828",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1016/j.antiviral.2016.11.011",
"doi-asserted-by": "publisher",
"key": "ref38"
},
{
"DOI": "10.3390/v10040149",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"DOI": "10.1016/j.antiviral.2018.07.010",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"DOI": "10.1016/j.antiviral.2017.12.010",
"doi-asserted-by": "publisher",
"key": "ref41"
},
{
"DOI": "10.3389/fphar.2021.652335",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.2174/1389201022666210322124348",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"DOI": "10.3390/v10110592",
"doi-asserted-by": "publisher",
"key": "ref44"
},
{
"DOI": "10.1016/j.antiviral.2021.105012",
"doi-asserted-by": "publisher",
"key": "ref45"
},
{
"DOI": "10.1177/1934578X211016610",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1016/j.antiviral.2016.12.014",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.4308/hjb.22.2.67",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1038/srep14179",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.2174/157340611795564268",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.3390/molecules25215037",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"DOI": "10.1021/acs.jnatprod.0c01324",
"doi-asserted-by": "publisher",
"key": "ref52"
},
{
"DOI": "10.1080/07391102.2020.1760136",
"doi-asserted-by": "publisher",
"key": "ref53"
},
{
"DOI": "10.1111/j.1541-4337.2003.tb00016.x",
"doi-asserted-by": "publisher",
"key": "ref54"
},
{
"DOI": "10.1186/s12917-019-2067-6",
"doi-asserted-by": "publisher",
"key": "ref55"
},
{
"DOI": "10.7324/JAPS.2013.31209",
"doi-asserted-by": "publisher",
"key": "ref56"
},
{
"DOI": "10.1186/s43088-021-00138-3",
"doi-asserted-by": "publisher",
"key": "ref57"
},
{
"DOI": "10.1016/j.ddtec.2006.09.004",
"doi-asserted-by": "publisher",
"key": "ref58"
},
{
"DOI": "10.1124/pr.112.007336",
"doi-asserted-by": "publisher",
"key": "ref59"
},
{
"DOI": "10.21037/apm-21-3373",
"doi-asserted-by": "publisher",
"key": "ref60"
},
{
"DOI": "10.1016/j.clnesp.2021.10.016",
"doi-asserted-by": "publisher",
"key": "ref61"
},
{
"DOI": "10.1128/JVI.00809-18",
"doi-asserted-by": "publisher",
"key": "ref62"
},
{
"DOI": "10.1038/ja.2011.21",
"doi-asserted-by": "publisher",
"key": "ref63"
},
{
"DOI": "10.3390/md19020094",
"doi-asserted-by": "publisher",
"key": "ref64"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "publisher",
"key": "ref65"
},
{
"DOI": "10.1016/j.str.2020.06.006",
"doi-asserted-by": "publisher",
"key": "ref66"
},
{
"DOI": "10.1002/prot.25219",
"doi-asserted-by": "publisher",
"key": "ref67"
},
{
"DOI": "10.1007/s40203-021-00103-z",
"doi-asserted-by": "publisher",
"key": "ref68"
},
{
"DOI": "10.1093/nar/gkh354",
"doi-asserted-by": "publisher",
"key": "ref69"
},
{
"DOI": "10.1038/s41598-018-23982-3",
"doi-asserted-by": "publisher",
"key": "ref70"
}
],
"reference-count": 70,
"references-count": 70,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/23/19/11131"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Inorganic Chemistry",
"Organic Chemistry",
"Physical and Theoretical Chemistry",
"Computer Science Applications",
"Spectroscopy",
"Molecular Biology",
"General Medicine",
"Catalysis"
],
"subtitle": [],
"title": "Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus",
"type": "journal-article",
"volume": "23"
}