Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach
et al., Biointerface Research in Applied Chemistry, doi:10.33263/BRIAC132.155, Mar 2022
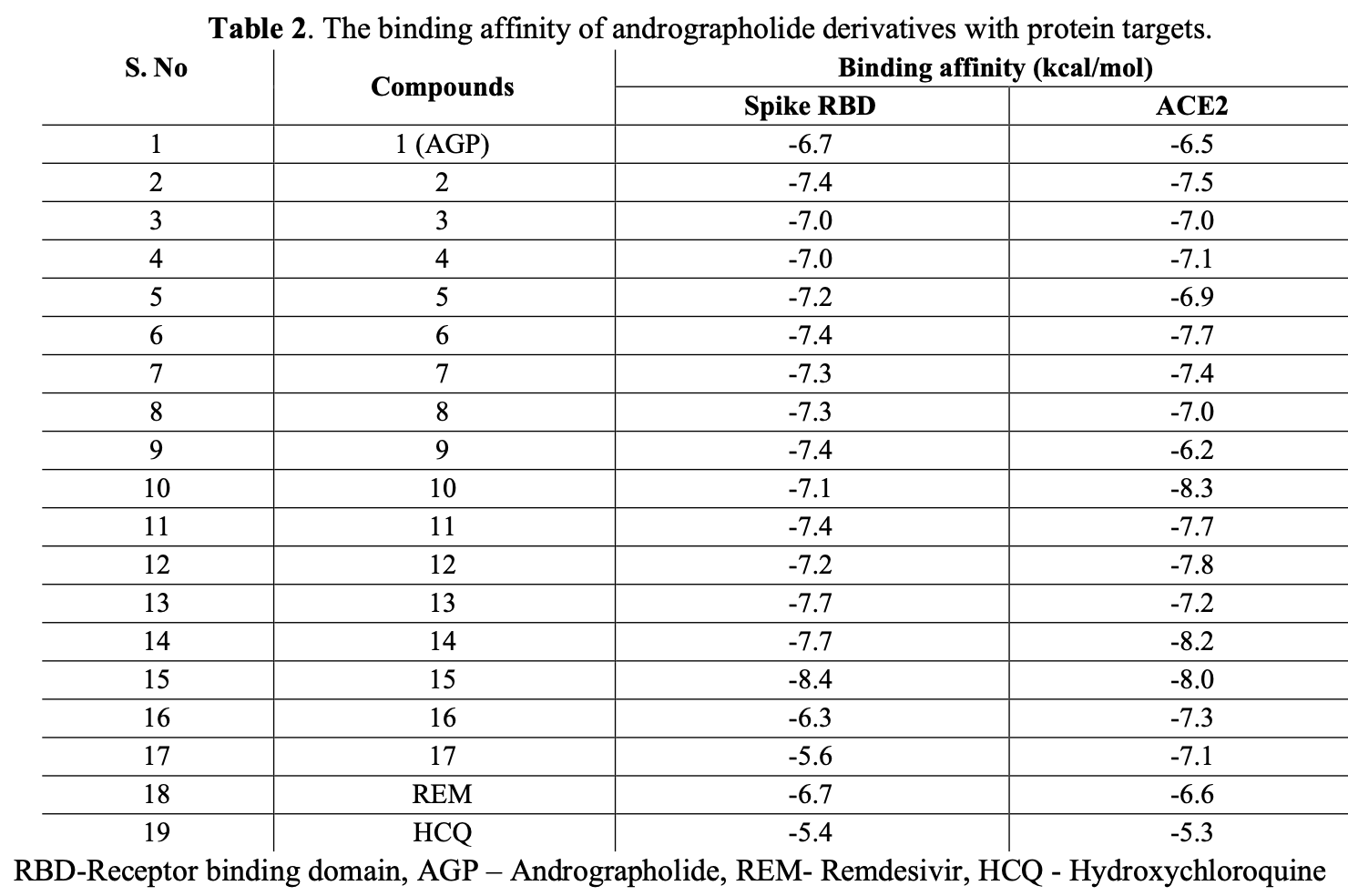
In silico study showing that semisynthetic andrographolide derivatives have potential to inhibit SARS-CoV-2 infection by targeting the spike protein and human ACE2 receptor. Docking studies found that derivatives AGP15 and AGP14 had the highest binding affinity for the spike protein (-8.4 and -7.7 kcal/mol respectively) while AGP10 and AGP14 had the highest binding affinity for the ACE2 receptor (-8.3 and -8.2 kcal/mol). The key spike protein residues involved in interactions were Arg355, Arg357, and Phe464, while the key ACE2 residues were Pro135, Glu160, Trp163, Lys689, and Asn690. Authors conclude AGP15 and AGP14 are promising anti-COVID-19 drug candidates that warrant further study.
25 preclinical studies support the efficacy of andrographolide for COVID-19:
In vitro studies demonstrate inhibition of the MproA,18 protein.
In vitro studies demonstrate efficacy in Calu-3B,18, A549C,14, and HUVECD,18 cells.
Animal studies demonstrate efficacy in Sprague Dawley miceE,18 and Golden Syrian hamstersF,14.
Andrographolide inhibits Mpro in a dose-dependent manner18, reduces ACE2 levels in the lung tissue of mice in combination with baicalein18, inhibits binding between the SARS-CoV-2 spike protein and ACE218, alleviates lung inflammation and cytokine storm in mice18, and improves survival and reduces lung inflammation via anti-inflammatory effects in Syrian hamsters14.
1.
Zhang et al., Effects and Mechanisms of Andrographolide for COVID-19: A Network Pharmacology-Based and Experimentally Validated Study, Natural Product Communications, doi:10.1177/1934578X241288428.
2.
Thomas et al., Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-58532-7.
3.
Arifin et al., Computational exploration of Andrographis paniculata herb compounds as potential antiviral agents targeting NSP3 (6W02) and NSP5 (7AR6) of SARS-COV-2, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2023.25.2.0292.
4.
Bhattarai et al., Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2174596.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
6.
Dassanayake et al., Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus, International Journal of Molecular Sciences, doi:10.3390/ijms231911131.
7.
Ningrum et al., Potency Of Andrographolide, L-Mimosine And Asiaticoside Compound As Antiviral For Covid-19 Based On In Silico Method, Proceedings Universitas Muhammadiyah Yogyakarta Undergraduate Conference, doi:10.18196/umygrace.v2i2.418.
8.
Ravichandran et al., Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach, Biointerface Research in Applied Chemistry, doi:10.33263/BRIAC132.155.
9.
Saeheng et al., In Silico Prediction of Andrographolide Dosage Regimens for COVID-19 Treatment, The American Journal of Chinese Medicine, doi:10.1142/S0192415X22500732.
10.
Khanal et al., Combination of system biology to probe the anti-viral activity of andrographolide and its derivative against COVID-19, RSC Advances, doi:10.1039/D0RA10529E.
11.
Rehan et al., A Computational Approach Identified Andrographolide as a Potential Drug for Suppressing COVID-19-Induced Cytokine Storm, Frontiers in Immunology, doi:10.3389/fimmu.2021.648250.
12.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
13.
Dey et al., The role of andrographolide and its derivative in COVID-19 associated proteins and immune system, Research Square, doi:10.21203/rs.3.rs-35800/v1.
14.
Kongsomros et al., In vivo evaluation of Andrographis paniculata and Boesenbergia rotunda extract activity against SARS-CoV-2 Delta variant in Golden Syrian hamsters: Potential herbal alternative for COVID-19 treatment, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2024.05.004.
15.
Chaopreecha et al., Andrographolide attenuates SARS-CoV-2 infection via an up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC), Phytomedicine, doi:10.1016/j.phymed.2024.156279.
16.
Li et al., Andrographolide suppresses SARS-CoV-2 infection by downregulating ACE2 expression: A mechanistic study, Antiviral Therapy, doi:10.1177/13596535241259952.
17.
Low et al., The wide spectrum anti-inflammatory activity of andrographolide in comparison to NSAIDs: a promising therapeutic compound against the cytokine storm, bioRxiv, doi:10.1101/2024.02.21.581396.
18.
Wan et al., Synergistic inhibition effects of andrographolide and baicalin on coronavirus mechanisms by downregulation of ACE2 protein level, Scientific Reports, doi:10.1038/s41598-024-54722-5.
19.
Siridechakorn et al., Inhibitory efficiency of Andrographis paniculata extract on viral multiplication and nitric oxide production, Scientific Reports, doi:10.1038/s41598-023-46249-y.
a.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
b.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
c.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
d.
HUVEC (Human Umbilical Vein Endothelial Cells) are primary endothelial cells derived from the vein of the umbilical cord. They are used to study vascular biology, including inflammation, angiogenesis, and viral interactions with endothelial cells.
e.
An outbred multipurpose breed of albino mouse used extensively in medical research.
f.
A rodent model widely used in infectious disease research due to their susceptibility to viral infections and similar disease progression to humans.
Ravichandran et al., 28 Mar 2022, Malaysia, peer-reviewed, 2 authors.
Contact: ravichandran_v@aimst.edu.my (corresponding author).
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach
Biointerface Research in Applied Chemistry, doi:10.33263/briac132.155
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causal factor for the current deadly infectious disease CoVID-19. There is no specific drug available for treating COVID-19 other than some vaccines approved for prevention. However, a lot of research is in progress to prove the anti-COVID-19 potential of natural and synthetic compounds. Objective: The present study was aimed to identify the anti-COVID-19 potential of andrographolide (AGP) derivatives by in-silico molecular interaction study. Seventeen AGP derivatives were screened for drug-likeness, ADME, and toxicity profile using in-silico online tools. Then the filtered AGP were subjected to molecular docking using the PyRx tool integrated with AutoDock Vina software. Compounds AGP 15, 14, and 10 have been identified as promising binding molecules for both S and ACE2, preventing the interaction between S and ACE2. AGP-15 had shown a -8.4 Kcal/mol binding/docking score for S, AGP-10 and 14 showed a -8.3 and -8.2 Kcal/mol binding/docking score for ACE2. Overall results indicated that AGP derivatives 15 and 14 might be the best candidates to battle COVID-19. However, further studies like dynamic molecular studies and pharmacological screenings are essential to confirm the stability and action potential of AGP derivatives 14 and 15 as a lead against COVID-19.
Conflicts of Interest The authors declare no conflict of interest.
References
Abdalla, Mohapatra, Sarangi, Mohapatra, Eltayb et al., In Silico Studies on Phytochemicals to Combat the Emerging COVID-19 Infection, J Saudi Chem Soc, doi:10.1016/j.jscs.2021.101367
Adeniji, Arthur, Oluwaseye, Computational Modeling of 4-Phenoxynicotinamide and 4-Phenoxypyrimidine-5-Carboxamide Derivatives as Potent Anti-Diabetic Agent against TGR5 Receptor, J King Saud Univ Sci, doi:10.1016/j.jksus.2018.03.007
Al-Shar'i, Tackling COVID-19: Identification of Potential Main Protease Inhibitors via Structural Analysis, Virtual Screening, Molecular Docking and MM-PBSA Calculations, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1800514
Alazmi, Motwalli, Molecular Basis for Drug Repurposing to Study the Interface of the S Protein in SARS-CoV-2 and Human ACE2 through Docking, Characterization, and Molecular Dynamics for Natural Drug Candidates, J Mol Model, doi:10.1007/s00894-020-04599-8
Cai, Zeng, Chen, The Pharmacological Mechanism of Huashi Baidu Formula for the Treatment of COVID-19 by Combined Network Pharmacology and Molecular Docking, Ann Palliat Med, doi:10.21037/apm-20-1759
Cardoso, Giacomello, Rocha De Oliveira, Da Silva, De et al., A Combined Molecular Docking and Density Functional Theory Nuclear Magnetic Resonance Study of Trans-Dehydrocrotonin Interacting with COVID-19 Main Protease and Severe Acute Respiratory Syndrome Coronavirus 2 3C-Like Protease, J Nanosci Nanotechnol, doi:10.1166/jnn.2021.19475
Chiou, Hsu, Chen, Yang, Tsay et al., Repurposing Existing Drugs: Identification of SARS-CoV-2 3C-like Protease Inhibitors, J Enzyme Inhib Med Chem, doi:10.1080/14756366.2020.1850710
Dallakyan, Olson, Small-Molecule Library Screening by Docking with PyRx, Methods Mol Biol, doi:10.1007/978-1-4939-2269-7_19
Dey, Low Bioavailability Hinders Drug Discovery against COVID-19, Guided by in Silico Docking, Br J Pharmacol, doi:10.1111/bph.15325
Elfiky, Anti-HCV, Nucleotide Inhibitors, Repurposing against COVID-19, Life Sci, doi:10.1016/j.lfs.2020.117477
Enayatkhani, Hasaniazad, Faezi, Gouklani, Davoodian et al., Reverse Vaccinology Approach to Design a Novel Multi-Epitope Vaccine Candidate against COVID-19: An in Silico Study, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1756411
Enmozhi, Raja, Sebastine, Joseph, Andrographolide as a Potential Inhibitor of SARS-CoV-2 Main Protease: An in Silico Approach, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1760136
Farhat, Khan, Repurposing Drug Molecule against SARS-Cov-2 (COVID-19) through Molecular Docking and Dynamics: A Quick Approach to Pick FDA-Approved Drugs, J Mol Model, doi:10.1007/s00894-021-04923-w
Gowrishankar, Muthumanickam, Kamaladevi, Karthika, Jothi et al., Promising Phytochemicals of Traditional Indian Herbal Steam Inhalation Therapy to Combat COVID-19-An in Silico Study, Food Chem Toxicol, doi:10.1016/j.fct.2020.111966
Gu, Zhang, Cen, Wu, Lu et al., Quercetin as a Potential Treatment for COVID-19-Induced Acute Kidney Injury: Based on Network Pharmacology and Molecular Docking Study, PLoS One, doi:10.1371/journal.pone.0245209
Gurung, Ali, Lee, Farah, Al-Anazi, Identification of Potential SARS-CoV-2 Entry Inhibitors by Targeting the Interface Region between the Spike RBD and Human ACE2, J Infec Public Health, doi:10.1016/j.jiph.2020.12.014
Ibrahim, Abdelrahman, Allemailem, Almatroudi, Moustafa et al., In Silico Evaluation of Prospective Anti-COVID-19 Drug Candidates as Potential SARS-CoV-2 Main Protease Inhibitors, Protein J, doi:10.1007/s10930-020-09945-6
Jayakumar, Hsieh, Lee, Sheu, Experimental and Clinical Pharmacology of Andrographis Paniculata and Its Major Bioactive Phytoconstituent Andrographolide, Evid Based Complement Alternat Med, doi:10.1155/2013/846740
Kanjanasirirat, Suksatu, Manopwisedjaroen, Munyoo, Tuchinda et al., High-Content Screening of Thai Medicinal Plants Reveals Boesenbergia Rotunda Extract and Its Component Panduratin A as Anti-SARS-CoV-2 Agents, Sci Rep, doi:10.1038/s41598-020-77003-3
Kodchakorn, Poovorawan, Suwannakarn, Kongtawelert, Molecular Modelling Investigation for Drugs and Nutraceuticals against Protease of SARS-CoV-2, J Mol Graph Model, doi:10.1016/j.jmgm.2020.107717
Kumar, Kashyap, Chowdhury, Kumar, Panwar et al., Identification of Phytochemicals as Potential Therapeutic Agents That Binds to Nsp15 Protein Target of Coronavirus (SARS-CoV-2) That Are Capable of Inhibiting Virus Replication, Phytomedicine, doi:10.1016/j.phymed.2020.153317
Kumar, Kumari, Vishvakarma, Jayaraj, Kumar et al., Promising Inhibitors of Main Protease of Novel Corona Virus to Prevent the Spread of COVID-19 Using Docking and Molecular Dynamics Simulation, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1779131
Lakshmi, Shafreen, Priya, Shunmugiah, Ethnomedicines of Indian Origin for Combating COVID-19 Infection by Hampering the Viral Replication: Using Structure-Based Drug Discovery Approach, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1778537
Li, He, Tang, Ding, Chu et al., Andrographolide Inhibits Inflammatory Cytokines Secretion in LPS-Stimulated RAW264. 7 Cells through Suppression of NF-ΚB/MAPK Signaling Pathway, Evid Based Complement Alternat Med, doi:10.1155/2017/8248142
Li, Li, Farzan, Harrison, Structure of SARS Coronavirus Spike Receptor-binding Domain Complexed with Receptor, Science, doi:10.1126/science.1116480
Li, Luo, Wang, Liao, Li, Inhibition of NF-KappaB Expression and Allergen-Induced Airway Inflammation in a Mouse Allergic Asthma Model by Andrographolide, Cell Mol Immunol, doi:10.1038/cmi.2009.49
Maurya, Kumar, Prasad, Bhatt, Saxena, Structure-Based Drug Designing for Potential Antiviral Activity of Selected Natural Products from Ayurveda against SARS-CoV-2 Spike Glycoprotein and Its Cellular Receptor, Virusdisease, doi:10.1007/s13337-020-00598-8
Muhammad, Maqbool, Al-Sehemi, Iqbal, Khan et al., A Threefold Approach Including Quantum Chemical, Molecular Docking and Molecular Dynamic Studies to Explore the Natural Compounds from Centaurea Jacea as the Potential Inhibitors for COVID-19, Braz J Biol, doi:10.1590/1519-6984.247604
Murugan, Pandian, Jeyakanthan, Computational Investigation on Andrographis Paniculata Phytochemicals to Evaluate Their Potency against SARS-CoV-2 in Comparison to Known Antiviral Compounds in Drug Trials, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1777901
Murugesan, Kottekad, Crasta, Sreevathsan, Usharani et al., Targeting COVID-19 (SARS-CoV-2) Main Protease through Active Phytocompounds of Ayurvedic Medicinal Plants -Emblica Officinalis (Amla), Phyllanthus Niruri Linn. (Bhumi Amla) and Tinospora Cordifolia (Giloy) -A Molecular Docking and Simulation Study, Comput Biol Med, doi:10.1016/j.compbiomed.2021.104683
Naik, Hule, Evaluation of Immunomodulatory Activity of an Extract of Andrographolides from Andographis Paniculata, Planta Med, doi:10.1055/s-0029-1185398
Rai, Barik, Singh, Suresh, Singh et al., Molecular Docking, Binding Mode Analysis, Molecular Dynamics, and Prediction of ADMET/Toxicity Properties of Selective Potential Antiviral Agents against SARS-CoV-2 Main Protease: An Effort toward Drug Repurposing to Combat COVID-19, Mol Divers, doi:10.1007/s11030-021-10188-5
Rajagopal, Varakumar, Aparna, Byran, Jupudi, Identification of Some Novel Oxazine Substituted 9-Anilinoacridines as SARS-CoV-2 Inhibitors for COVID-19 by Molecular Docking, Free Energy Calculation and Molecular Dynamics Studies, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1798285
Rathinavel, Thangaswamy, Ammashi, Kumarasamy, Virtual Screening of COVID-19 Drug from Three Indian Traditional Medicinal Plants through in Silico Approach, Res J Biotechnol
Sayed, Khalaf, Abdelrahim, Elgendy, Repurposing of Some Anti-Infective Drugs for COVID-19 Treatment: A Surveillance Study Supported by an in Silico Investigation, Int J Clin Pract, doi:10.1111/ijcp.13877
Shah, Bhaliya, Patel, In Silico Approach: Docking Study of Oxindole Derivatives against the Main Protease of COVID-19 and Its Comparison with Existing Therapeutic Agents, J Basic Clin Physiol Pharmacol, doi:10.1515/jbcpp-2020-0262
Sharma, Vora, Patel, Sinha, Jha et al., Identification of Natural Inhibitors against Prime Targets of SARS-CoV-2 Using Molecular Docking, Molecular Dynamics Simulation and MM-PBSA Approaches, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1846624
Shi, Huang, Chen, Pi, Hsu et al., Andrographolide and Its Fluorescent Derivative Inhibit the Main Proteases of 2019-NCoV and SARS-CoV through Covalent Linkage, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2020.08.086
Shin, Mukherjee, Grewe, Bojkova, Baek et al., Papainlike Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity, Nature, doi:10.1038/s41586-020-2601-5
Shree, Mishra, Selvaraj, Singh, Chaube et al., Targeting COVID-19 (SARS-CoV-2) Main Protease through Active Phytochemicals of Ayurvedic Medicinal Plants -Withania Somnifera (Ashwagandha), Tinospora Cordifolia (Giloy) and Ocimum Sanctum (Tulsi) -a Molecular Docking Study, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1810778
Sonkar, Doharey, Rathore, Singh, Kashyap et al., Repurposing of Gastric Cancer Drugs against COVID-19, Comput Biol Med, doi:10.1016/j.compbiomed.2021.104826
Stadler, Masignani, Eickmann, Becker, Abrignani et al., SARS-Beginning to Understand a New Virus, Nature Reviews Microbiology, doi:10.1038/nrmicro775
Sukardiman; Ervina, Fadhil Pratama, Poerwono, Siswodihardjo, The Coronavirus Disease 2019 Main Protease Inhibitor from Andrographis Paniculata (Burm. f) Ness, J Adv Pharm Technol Res, doi:10.4103/japtr.JAPTR_84_20
Suresh, Deepika, Bantal, Beedu, Rupula, Evaluation of Anti-Inflammatory and Anti-Nociceptive Potentials of Andrographolide and Forskolin: In Vivo Studies, J biol act prod nat, doi:10.1080/22311866.2018.1526650
Suručić, Tubić, Stojiljković, Djuric, Travar et al., Computational Study of Pomegranate Peel Extract Polyphenols as Potential Inhibitors of SARS-CoV-2 Virus Internalization, Mol Cell Biochem, doi:10.1007/s11010-020-03981-7
Vique-Sánchez, Potential Inhibitors Interacting in Neuropilin-1 to Develop an Adjuvant Drug against COVID-19, by Molecular Docking, Bioorg Med Chem, doi:10.1016/j.bmc.2021.116040
Wang, Liu, Zhang, Wilson, Hong, Andrographolide Reduces Inflammation-Mediated Dopaminergic Neurodegeneration in Mesencephalic Neuron-Glia Cultures by Inhibiting Microglial Activation, J Pharmacol Exp Ther, doi:10.1124/jpet.103.059683
Wang, Sun, Ding, The Therapeutic Effects of Traditional Chinese Medicine on COVID-19: A Narrative Review, Int J Clin Pharm, doi:10.1007/s11096-020-01153-7
Wang, Wang, Dong, Liu, Italiani et al., Immunomodulatory Activity of Andrographolide on Macrophage Activation and Specific Antibody Response, Acta Pharmacol Sin, doi:10.1038/aps.2009.205
Wipatayotin, Local Herb Study on Covid-19 Patients to Continue
Xu, Gao, Liang, Chen, In Silico Screening of Potential Anti-COVID-19 Bioactive Natural Constituents from Food Sources by Molecular Docking, Nutrition, doi:10.1016/j.nut.2020.111049
Çakır, Okuyan, Şener, Tunali-Akbay, Investigation of Beta-Lactoglobulin Derived Bioactive Peptides against SARS-CoV-2 (COVID-19): In Silico Analysis, Eur J Pharmacol, doi:10.1016/j.ejphar.2020.173781
DOI record:
{
"DOI": "10.33263/briac132.155",
"ISSN": [
"2069-5837"
],
"URL": "http://dx.doi.org/10.33263/BRIAC132.155",
"abstract": "<jats:p>Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causal factor for the current deadly infectious disease CoVID-19. There is no specific drug available for treating COVID-19 other than some vaccines approved for prevention. However, a lot of research is in progress to prove the anti-COVID-19 potential of natural and synthetic compounds. Objective: The present study was aimed to identify the anti-COVID-19 potential of andrographolide (AGP) derivatives by in-silico molecular interaction study. Seventeen AGP derivatives were screened for drug-likeness, ADME, and toxicity profile using in-silico online tools. Then the filtered AGP were subjected to molecular docking using the PyRx tool integrated with AutoDock Vina software. Compounds AGP 15, 14, and 10 have been identified as promising binding molecules for both S and ACE2, preventing the interaction between S and ACE2. AGP-15 had shown a −8.4 Kcal/mol binding/docking score for S, AGP-10 and 14 showed a -8.3 and -8.2 Kcal/mol binding/docking score for ACE2. Overall results indicated that AGP derivatives 15 and 14 might be the best candidates to battle COVID-19. However, further studies like dynamic molecular studies and pharmacological screenings are essential to confirm the stability and action potential of AGP derivatives 14 and 15 as a lead against COVID-19.</jats:p>",
"container-title": "Biointerface Research in Applied Chemistry",
"container-title-short": "Biointerface Res Appl Chem",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
4,
10
]
],
"date-time": "2022-04-10T06:52:03Z",
"timestamp": 1649573523000
},
"deposited": {
"date-parts": [
[
2022,
4,
10
]
],
"date-time": "2022-04-10T06:52:05Z",
"timestamp": 1649573525000
},
"indexed": {
"date-parts": [
[
2022,
4,
10
]
],
"date-time": "2022-04-10T07:10:33Z",
"timestamp": 1649574633186
},
"is-referenced-by-count": 0,
"issue": "2",
"issued": {
"date-parts": [
[
2022,
3,
28
]
]
},
"journal-issue": {
"issue": "2",
"published-online": {
"date-parts": [
[
2022,
4,
7
]
]
}
},
"language": "en",
"member": "18784",
"original-title": [],
"page": "155",
"prefix": "10.33263",
"published": {
"date-parts": [
[
2022,
3,
28
]
]
},
"published-online": {
"date-parts": [
[
2022,
3,
28
]
]
},
"publisher": "AMG Transcend Association",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://biointerfaceresearch.com/wp-content/uploads/2022/03/BRIAC132.155.pdf"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Molecular Biology",
"Molecular Medicine",
"Biochemistry",
"Biotechnology"
],
"subtitle": [],
"title": "Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach",
"type": "journal-article",
"volume": "13"
}