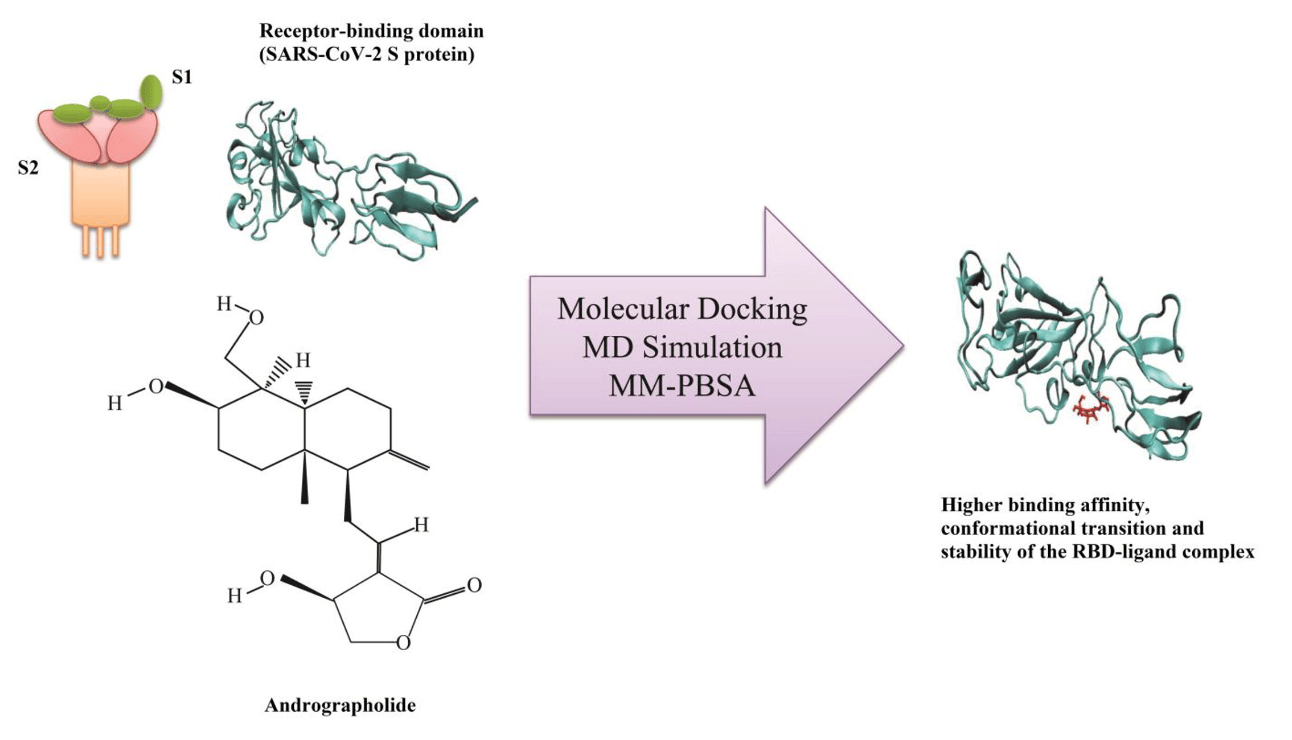
Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations
et al., Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2174596, Feb 2023
In silico study showing that andrographolide binds strongly to the SARS-CoV-2 spike protein receptor-binding domain (RBD). Authors used molecular docking and dynamics simulations to demonstrate that andrographolide forms stable non-covalent interactions with the RBD, leading to conformational changes that stabilize the complex. The ligand-bound RBD showed reduced conformational fluctuations compared to the unbound form, suggesting potential interference with ACE2 receptor binding and viral entry.
25 preclinical studies support the efficacy of andrographolide for COVID-19:
In vitro studies demonstrate inhibition of the MproA,18 protein.
In vitro studies demonstrate efficacy in Calu-3B,18, A549C,14, and HUVECD,18 cells.
Animal studies demonstrate efficacy in Sprague Dawley miceE,18 and Golden Syrian hamstersF,14.
Andrographolide inhibits Mpro in a dose-dependent manner18, reduces ACE2 levels in the lung tissue of mice in combination with baicalein18, inhibits binding between the SARS-CoV-2 spike protein and ACE218, alleviates lung inflammation and cytokine storm in mice18, and improves survival and reduces lung inflammation via anti-inflammatory effects in Syrian hamsters14.
1.
Zhang et al., Effects and Mechanisms of Andrographolide for COVID-19: A Network Pharmacology-Based and Experimentally Validated Study, Natural Product Communications, doi:10.1177/1934578X241288428.
2.
Thomas et al., Cheminformatics approach to identify andrographolide derivatives as dual inhibitors of methyltransferases (nsp14 and nsp16) of SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-024-58532-7.
3.
Arifin et al., Computational exploration of Andrographis paniculata herb compounds as potential antiviral agents targeting NSP3 (6W02) and NSP5 (7AR6) of SARS-COV-2, GSC Biological and Pharmaceutical Sciences, doi:10.30574/gscbps.2023.25.2.0292.
4.
Bhattarai et al., Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations, Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2023.2174596.
5.
Nguyen et al., The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics, Bioinformatics and Biology Insights, doi:10.1177/11779322221149622.
6.
Dassanayake et al., Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus, International Journal of Molecular Sciences, doi:10.3390/ijms231911131.
7.
Ningrum et al., Potency Of Andrographolide, L-Mimosine And Asiaticoside Compound As Antiviral For Covid-19 Based On In Silico Method, Proceedings Universitas Muhammadiyah Yogyakarta Undergraduate Conference, doi:10.18196/umygrace.v2i2.418.
8.
Ravichandran et al., Identification of Potential Semisynthetic Andrographolide Derivatives to Combat COVID-19 by Targeting the SARS-COV-2 Spike Protein and Human ACE2 Receptor– An In-silico Approach, Biointerface Research in Applied Chemistry, doi:10.33263/BRIAC132.155.
9.
Saeheng et al., In Silico Prediction of Andrographolide Dosage Regimens for COVID-19 Treatment, The American Journal of Chinese Medicine, doi:10.1142/S0192415X22500732.
10.
Khanal et al., Combination of system biology to probe the anti-viral activity of andrographolide and its derivative against COVID-19, RSC Advances, doi:10.1039/D0RA10529E.
11.
Rehan et al., A Computational Approach Identified Andrographolide as a Potential Drug for Suppressing COVID-19-Induced Cytokine Storm, Frontiers in Immunology, doi:10.3389/fimmu.2021.648250.
12.
Rajagopal et al., Activity of phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach, Future Journal of Pharmaceutical Sciences, doi:10.1186/s43094-020-00126-x.
13.
Dey et al., The role of andrographolide and its derivative in COVID-19 associated proteins and immune system, Research Square, doi:10.21203/rs.3.rs-35800/v1.
14.
Kongsomros et al., In vivo evaluation of Andrographis paniculata and Boesenbergia rotunda extract activity against SARS-CoV-2 Delta variant in Golden Syrian hamsters: Potential herbal alternative for COVID-19 treatment, Journal of Traditional and Complementary Medicine, doi:10.1016/j.jtcme.2024.05.004.
15.
Chaopreecha et al., Andrographolide attenuates SARS-CoV-2 infection via an up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC), Phytomedicine, doi:10.1016/j.phymed.2024.156279.
16.
Li et al., Andrographolide suppresses SARS-CoV-2 infection by downregulating ACE2 expression: A mechanistic study, Antiviral Therapy, doi:10.1177/13596535241259952.
17.
Low et al., The wide spectrum anti-inflammatory activity of andrographolide in comparison to NSAIDs: a promising therapeutic compound against the cytokine storm, bioRxiv, doi:10.1101/2024.02.21.581396.
18.
Wan et al., Synergistic inhibition effects of andrographolide and baicalin on coronavirus mechanisms by downregulation of ACE2 protein level, Scientific Reports, doi:10.1038/s41598-024-54722-5.
19.
Siridechakorn et al., Inhibitory efficiency of Andrographis paniculata extract on viral multiplication and nitric oxide production, Scientific Reports, doi:10.1038/s41598-023-46249-y.
a.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
b.
Calu-3 is a human lung adenocarcinoma cell line with moderate ACE2 and TMPRSS2 expression and SARS-CoV-2 susceptibility. It provides a model of the human respiratory epithelium, but many not be ideal for modeling early stages of infection due to the moderate expression levels of ACE2 and TMPRSS2.
c.
A549 is a human lung carcinoma cell line with low ACE2 expression and SARS-CoV-2 susceptibility. Viral entry/replication can be studied but the cells may not replicate all aspects of lung infection.
d.
HUVEC (Human Umbilical Vein Endothelial Cells) are primary endothelial cells derived from the vein of the umbilical cord. They are used to study vascular biology, including inflammation, angiogenesis, and viral interactions with endothelial cells.
e.
An outbred multipurpose breed of albino mouse used extensively in medical research.
f.
A rodent model widely used in infectious disease research due to their susceptibility to viral infections and similar disease progression to humans.
Bhattarai et al., 10 Feb 2023, peer-reviewed, 3 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
DOI record:
{
"DOI": "10.1080/07391102.2023.2174596",
"ISSN": [
"0739-1102",
"1538-0254"
],
"URL": "http://dx.doi.org/10.1080/07391102.2023.2174596",
"alternative-id": [
"10.1080/07391102.2023.2174596"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=tbsd20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=tbsd20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2022-09-19"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2023-01-24"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2023-02-10"
}
],
"author": [
{
"affiliation": [
{
"name": "Bioinformatics Programming Laboratory, Department of Biotechnology, School of Bio-Sciences and Technology, Vellore Institute of Technology, VIT, Vellore, Tamil Nadu, India"
}
],
"family": "Bhattarai",
"given": "Anil",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Bioinformatics Programming Laboratory, Department of Biotechnology, School of Bio-Sciences and Technology, Vellore Institute of Technology, VIT, Vellore, Tamil Nadu, India"
}
],
"family": "Priyadharshini",
"given": "Annadurai",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4212-0927",
"affiliation": [
{
"name": "Bioinformatics Programming Laboratory, Department of Biotechnology, School of Bio-Sciences and Technology, Vellore Institute of Technology, VIT, Vellore, Tamil Nadu, India"
}
],
"authenticated-orcid": false,
"family": "Emerson",
"given": "Isaac Arnold",
"sequence": "additional"
}
],
"container-title": "Journal of Biomolecular Structure and Dynamics",
"container-title-short": "Journal of Biomolecular Structure and Dynamics",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2023,
2,
11
]
],
"date-time": "2023-02-11T02:20:36Z",
"timestamp": 1676082036000
},
"deposited": {
"date-parts": [
[
2024,
3,
5
]
],
"date-time": "2024-03-05T18:37:15Z",
"timestamp": 1709663835000
},
"indexed": {
"date-parts": [
[
2024,
3,
5
]
],
"date-time": "2024-03-05T19:10:40Z",
"timestamp": 1709665840476
},
"is-referenced-by-count": 2,
"issue": "22",
"issued": {
"date-parts": [
[
2023,
2,
10
]
]
},
"journal-issue": {
"issue": "22",
"published-print": {
"date-parts": [
[
2023,
12,
29
]
]
}
},
"language": "en",
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/07391102.2023.2174596",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "13438-13453",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2023,
2,
10
]
]
},
"published-online": {
"date-parts": [
[
2023,
2,
10
]
]
},
"published-print": {
"date-parts": [
[
2023,
12,
29
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.3109/14756366.2014.979345",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_2_1"
},
{
"DOI": "10.1016/j.softx.2015.06.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_1"
},
{
"DOI": "10.1007/s00894-020-04599-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_4_1"
},
{
"DOI": "10.1080/07391102.2021.1900916",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_1"
},
{
"DOI": "10.1371/journal.pcbi.1000544",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_1"
},
{
"DOI": "10.1093/nar/gky318",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_1"
},
{
"DOI": "10.1080/07391102.2018.1444511",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_1"
},
{
"DOI": "10.1021/j100142a004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_9_1"
},
{
"DOI": "10.3389/fimmu.2020.552925",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_1"
},
{
"DOI": "10.1002/prot.22922",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_1"
},
{
"DOI": "10.1128/JVI.77.16.8801-8811.2003",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_12_1"
},
{
"DOI": "10.1093/bioinformatics/btx349",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_13_1"
},
{
"DOI": "10.1080/22221751.2020.1725399",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_1"
},
{
"DOI": "10.1080/22221751.2020.1737364",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_1"
},
{
"DOI": "10.1038/s41579-018-0118-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_1"
},
{
"DOI": "10.1016/j.toxicon.2021.02.015",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_1"
},
{
"DOI": "10.1063/1.464397",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_1"
},
{
"DOI": "10.1021/acs.jmedchem.5b01684",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_1"
},
{
"DOI": "10.1093/nar/gku401",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_1"
},
{
"DOI": "10.1007/978-1-4939-2438-7_1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_1"
},
{
"DOI": "10.1016/0040-4020(80)80168-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_1"
},
{
"DOI": "10.1002/elps.1150181505",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_1"
},
{
"article-title": "Avogadro: An advanced semantic chemical editor, visualization, and analysis platform",
"author": "Hanwell M. D.",
"journal-title": "Journal of Cheminformatics 4(1), 1-17.",
"key": "e_1_3_3_24_1",
"unstructured": "Hanwell, M. D., Curtis, D. E., Lonie, D. C., Vandermeersch, T., Zurek, E., & Hutchison, G. R. (2018). Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics 4(1), 1-17.",
"year": "2018"
},
{
"DOI": "10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_1"
},
{
"DOI": "10.1038/s41579-020-00459-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_1"
},
{
"DOI": "10.1002/jcc.23354",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_27_1"
},
{
"DOI": "10.1038/nmeth.4067",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_1"
},
{
"DOI": "10.1107/S090744490200985X",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_1"
},
{
"DOI": "10.1002/prot.340110305",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_1"
},
{
"DOI": "10.1103/PhysRevLett.94.078102",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_1"
},
{
"DOI": "10.1155/2013/846740",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_1"
},
{
"DOI": "10.1038/189771a0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_1"
},
{
"DOI": "10.1038/nsb0902-646",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_1"
},
{
"DOI": "10.1038/347631a0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_35_1"
},
{
"DOI": "10.1021/ci500020m",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_36_1"
},
{
"article-title": "Influence of logBB cut-off on the prediction of blood-brain barrier permeability",
"author": "Kunwittaya S.",
"first-page": "16",
"journal-title": "Biomedical and Applied Technology Journal, 1",
"key": "e_1_3_3_37_1",
"unstructured": "Kunwittaya, S., Nantasenamat, C., Treeratanapiboon, L., Srisarin, A., Isarankura-Na-Ayudhya, C., & Prachayasittikul, V. (2013). Influence of logBB cut-off on the prediction of blood-brain barrier permeability. Biomedical and Applied Technology Journal, 1, 16–34.",
"year": "2013"
},
{
"DOI": "10.1016/S1875-5364(20)60016-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_38_1"
},
{
"DOI": "10.3390/ph13090209",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_39_1"
},
{
"DOI": "10.1038/s41564-020-0688-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_40_1"
},
{
"DOI": "10.1002/prot.22711",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_41_1"
},
{
"DOI": "10.1016/j.ddtec.2004.11.007",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_42_1"
},
{
"DOI": "10.15171/apb.2017.041",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_43_1"
},
{
"DOI": "10.1016/j.str.2011.03.010",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_44_1"
},
{
"article-title": "Andrographis paniculata (Kalmegh). A Review",
"author": "Mishra S. K.",
"first-page": "283",
"journal-title": "Pharmacognosy Reviews, 1",
"key": "e_1_3_3_45_1",
"unstructured": "Mishra, S. K., Sangwan, N. S., & Sangwan, R. S. (2007). Andrographis paniculata (Kalmegh). A Review. Pharmacognosy Reviews, 1(2), 283–298.",
"year": "2007"
},
{
"DOI": "10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_46_1"
},
{
"DOI": "10.1002/cbic.202000047",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_47_1"
},
{
"DOI": "10.3390/v13050828",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_48_1"
},
{
"DOI": "10.1016/j.chempr.2021.07.015",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_49_1"
},
{
"DOI": "10.1016/j.tibs.2021.06.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_50_1"
},
{
"DOI": "10.1021/jp5013544",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_51_1"
},
{
"DOI": "10.1021/acsomega.2c00844",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_52_1"
},
{
"DOI": "10.1007/s15010-020-01486-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_53_1"
},
{
"DOI": "10.1021/acs.jnatprod.0c01324",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_54_1"
},
{
"DOI": "10.1021/jacs.0c01718",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_55_1"
},
{
"DOI": "10.3389/fmolb.2020.599079",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_56_1"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_57_1"
},
{
"DOI": "10.1163/_q3_SIM_00374",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_58_1"
},
{
"DOI": "10.1002/jcc.20035",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_59_1"
},
{
"DOI": "10.1038/s41401-021-00735-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_60_1"
},
{
"DOI": "10.1016/j.cell.2020.03.045",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_61_1"
},
{
"DOI": "10.1002/cpbi.3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_62_1"
},
{
"DOI": "10.1016/j.apsb.2021.06.016",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_63_1"
},
{
"DOI": "10.1002/jcc.21816",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_64_1"
}
],
"reference-count": 63,
"references-count": 63,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/07391102.2023.2174596"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Investigating the binding affinity of andrographolide against human SARS-CoV-2 spike receptor-binding domain through docking and molecular dynamics simulations",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1080/tandf_crossmark_01",
"volume": "41"
}