Within-host dynamics of antiviral treatment with remdesivir for SARS-CoV-2 infection
et al., Journal of The Royal Society Interface, doi:10.1098/rsif.2024.0536, Nov 2024
Retrospective 369 COVID-19 patients in Greece showing no significant improvement in mortality with remdesivir treatment. The study tracked 88 patients in detail, including 19 treated with remdesivir, and found that delayed hospitalization correlated with higher mortality among treated patients, however remdesivir showed limited antiviral effectiveness and no impact on survival.
Gérard, Zhou, Wu, Kamo, Choi, Kim show increased risk of acute kidney injury, Leo, Briciu, Muntean, Petrov show increased risk of liver injury, and Negru, Cheng, Mohammed, Kwok show increased risk of cardiac disorders with remdesivir.
1.
Gérard et al., Remdesivir and Acute Renal Failure: A Potential Safety Signal From Disproportionality Analysis of the WHO Safety Database, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.2145.
2.
Zhou et al., Acute Kidney Injury and Drugs Prescribed for COVID-19 in Diabetes Patients: A Real-World Disproportionality Analysis, Frontiers in Pharmacology, doi:10.3389/fphar.2022.833679.
3.
Wu et al., Acute Kidney Injury Associated With Remdesivir: A Comprehensive Pharmacovigilance Analysis of COVID-19 Reports in FAERS, Frontiers in Pharmacology, doi:10.3389/fphar.2022.692828.
4.
Kamo et al., Association of Antiviral Drugs for the Treatment of COVID-19 With Acute Renal Failure, In Vivo, doi:10.21873/invivo.13637.
5.
Choi et al., Comparative effectiveness of combination therapy with nirmatrelvir–ritonavir and remdesivir versus monotherapy with remdesivir or nirmatrelvir–ritonavir in patients hospitalised with COVID-19: a target trial emulation study, The Lancet Infectious Diseases, doi:10.1016/S1473-3099(24)00353-0.
6.
Kim et al., Investigating the Safety Profile of Fast‐Track COVID‐19 Drugs Using the FDA Adverse Event Reporting System Database: A Comparative Observational Study, Pharmacoepidemiology and Drug Safety, doi:10.1002/pds.70043.
7.
Leo et al., Hepatocellular liver injury in hospitalized patients affected by COVID-19: Presence of different risk factors at different time points, Digestive and Liver Disease, doi:10.1016/j.dld.2021.12.014.
8.
Briciu et al., Evolving Clinical Manifestations and Outcomes in COVID-19 Patients: A Comparative Analysis of SARS-CoV-2 Variant Waves in a Romanian Hospital Setting, Pathogens, doi:10.3390/pathogens12121453.
9.
Muntean et al., Effects of COVID-19 on the Liver and Mortality in Patients with SARS-CoV-2 Pneumonia Caused by Delta and Non-Delta Variants: An Analysis in a Single Centre, Pharmaceuticals, doi:10.3390/ph17010003.
10.
Petrov et al., The Effect of Potentially Hepatotoxic Medicinal Products on Alanine Transaminase Levels in COVID-19 Patients: A Case–Control Study, Safety and Risk of Pharmacotherapy, doi:10.30895/2312-7821-2025-458.
11.
Negru et al., Comparative Pharmacovigilance Analysis of Approved and Repurposed Antivirals for COVID-19: Insights from EudraVigilance Data, Biomedicines, doi:10.3390/biomedicines13061387.
12.
Cheng et al., Cardiovascular Safety of COVID-19 Treatments: A Disproportionality Analysis of Adverse Event Reports from the WHO VigiBase, Infectious Diseases and Therapy, doi:10.1007/s40121-025-01225-z.
Schuh et al., 27 Nov 2024, retrospective, Greece, peer-reviewed, 7 authors.
Within-host dynamics of antiviral treatment with remdesivir for SARS-CoV-2 infection
Journal of The Royal Society Interface, doi:10.1098/rsif.2024.0536
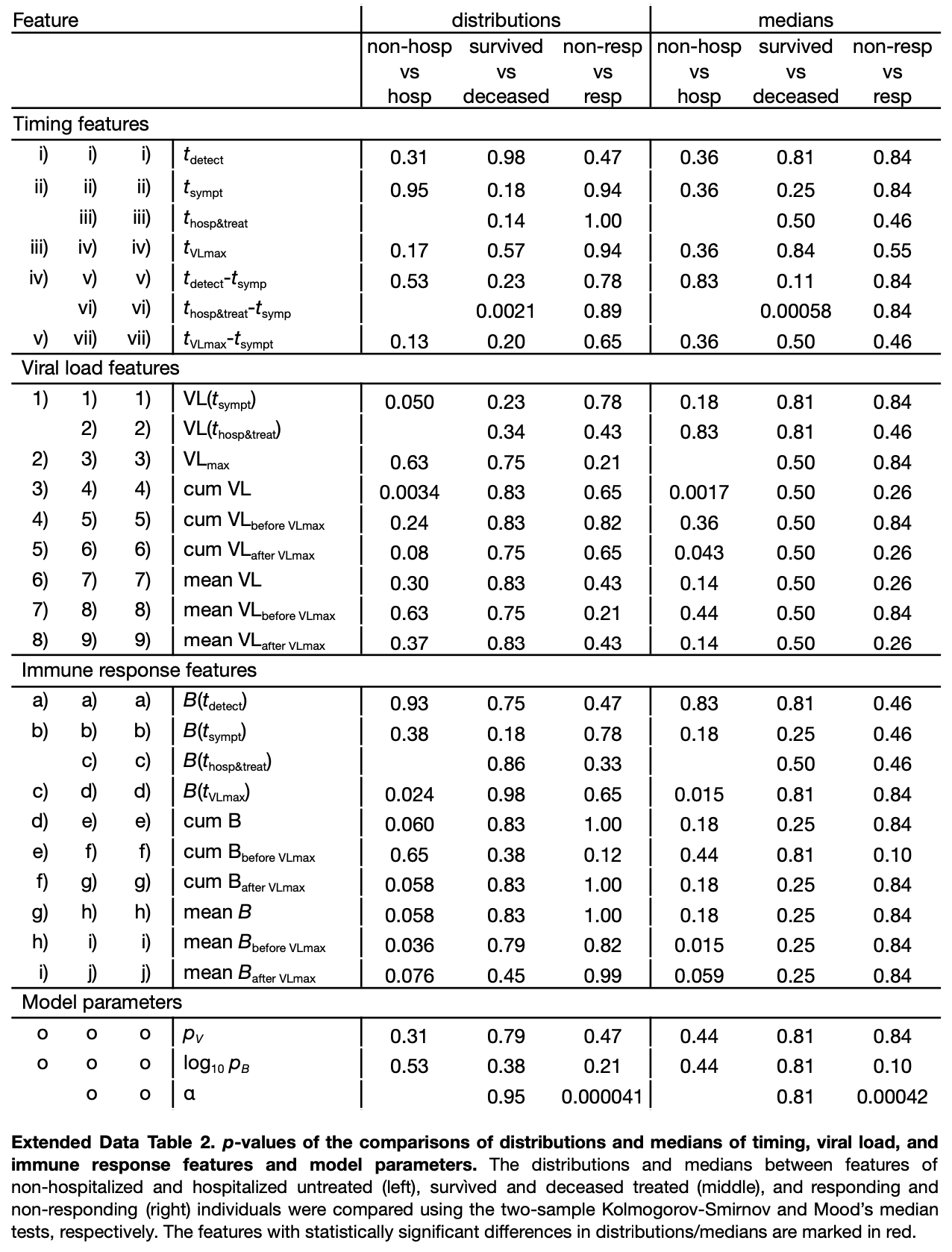
The effectiveness of antiviral treatment with remdesivir against COVID-19 has been investigated in clinical trials suggesting earlier recovery. However, this effect seems to be rather modest. In this study, we tracked the clinical course of SARS-CoV-2 infections in 369 COVID-19 individuals across a spectrum of illness severities, including both untreated individuals and individuals who received antiviral treatment with remdesivir. Moreover, using a process-based mathematical model, we quantified and analysed the within-host infection dynamics of a total of 88 individuals, of which 69 were untreated and 19 antiviral-treated individuals. For untreated individuals, we found that those hospitalized exhibit lower levels of early immune response and higher cumulative viral loads than those who were not. For treated individuals, we found that those who died were on average hospitalized later after symptom onset than those who survived, underscoring the importance of early medical intervention for severe COVID-19. Finally, our model estimates a rather limited antiviral activity of remdesivir. Our results provide valuable insights into the clinical course of COVID-19 during antiviral treatment with remdesivir and suggest the need for alternative treatment regimens.
Ethics. The research protocol was approved by the Research Ethics Committee of the Faculty of Medicine, University of Thessaly, Greece (84/09.12.2022) and was also approved by the Scientific Council of the public hospital from which the sample was derived (19535/28.06.2023). Data accessibility. The MATLAB code corresponding to this manuscript can be found via a permanent repository on Zenodo [44] . Some restrictions apply to the clinical and epidemiological data and are available to interested researchers upon request to the Research Ethics Committee of the University of Thessaly (contact: ehde@uth.gr). Supplementary material is available online [45] . Declaration of AI use. Conflict of interest declaration. We declare we have no competing interests.
References
Ader, Bouscambert-Duchamp, Hites, Peiffer-Smadja, Mentré et al., Final results of the DisCoVeRy trial of remdesivir for patients admitted to hospital with COVID-19, Lancet Infect. Dis, doi:10.1016/S1473-3099(22)00295-X
Adéoti, Agbla, Diop, Kakaï, Nonlinear mixed models and related approaches in infectious disease modeling: a systematic and critical review, Infect. Dis. Model, doi:10.1016/j.idm.2024.09.001
Barratt-Due, Evaluation of the Effects of Remdesivir and hydroxychloroquine on viral clearance in COVID-19 : a randomized trial, Ann. Intern. Med, doi:10.7326/M21-0653
Beigel, Remdesivir for the treatment of Covid-19 -final report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Biancofiore, Mirijello, Puteo, Viesti, Labonia et al., Remdesivir significantly reduces SARS-CoV-2 viral load on nasopharyngeal swabs in hospitalized patients with COVID-19: a retrospective case-control study, J. Med. Virol, doi:10.1002/jmv.27598
Campogiani, Remdesivir influence on SARS-CoV-2 RNA viral load kinetics in nasopharyngeal swab specimens of COVID-19 hospitalized patients: a real-life experience, Microorganisms, doi:10.3390/microorganisms11020312
Challenger, Foo, Wu, Yan, Marjaneh et al., Modelling upper respiratory viral load dynamics of SARS-CoV-2, BMC Med, doi:10.1186/s12916-021-02220-0
Elias, Sekri, Leblanc, Cucherat, Vanhems, The incubation period of COVID-19: a meta-analysis, Int. J. Infect. Dis, doi:10.1016/j.ijid.2021.01.069
Ferretti, The timing of COVID-19 transmission, SSRN Journal, doi:10.2139/ssrn.3716879
Goldberg, Zvi, Sheena, Sofer, Krause et al., A real-life setting evaluation of the effect of remdesivir on viral load in COVID-19 patients admitted to a large tertiary centre in Israel, Clin. Microbiol. Infect, doi:10.1016/j.cmi.2021.02.029
Goyal, Duke, Cardozo-Ojeda, Schiffer, Modeling explains prolonged SARS-CoV-2 nasal shedding relative to lung shedding in remdesivir-treated rhesus macaques, i. Sci, doi:10.1016/j.isci.2022.104448
Hass, Loos, Raimúndez-Álvarez, Timmer, Hasenauer et al., Benchmark problems for dynamic modeling of intracellular processes, Bioinformatics, doi:10.1093/bioinformatics/btz020
Kass, Raftery, Bayes factors, J. Am. Stat. Assoc, doi:10.1080/01621459.1995.10476572
Ke, Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness, Nat. Microbiol, doi:10.1038/s41564-022-01105-z
Ke, Zitzmann, Ho, Ribeiro, Perelson, In vivo kinetics of SARS-CoV-2 infection and its relationship with a person's infectiousness, Proc. Natl Acad. Sci. USA, doi:10.1073/pnas.2111477118
Keine, Mood's median test
Kokic, Mechanism of SARS-CoV-2 polymerase stalling by remdesivir, Nat. Commun, doi:10.1038/s41467-020-20542-0
Lingas, Effect of remdesivir on viral dynamics in COVID-19 hospitalized patients: a modelling analysis of the randomized, controlled, open-label DisCoVeRy trial, J. Antimicrob. Chemother, doi:10.1093/jac/dkac048
Marc, Impact of variants of concern on SARS-CoV-2 viral dynamics in non-human primates, PLoS Comput. Biol, doi:10.1371/journal.pcbi.1010721
Néant, Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort, Proc. Natl Acad. Sci. USA, doi:10.1073/pnas.2017962118
Owens, Esmaeili, Schiffer, Heterogeneous SARS-CoV-2 kinetics due to variable timing and intensity of immune responses, JCI Insight, doi:10.1172/jci.insight.176286
Pan, Repurposed antiviral drugs for Covid-19 -interim WHO Solidarity Trial results, N. Engl. J. Med, doi:10.1056/NEJMoa2023184
Perelson, Ke, Mechanistic modeling of SARS-CoV-2 and other infectious diseases and the effects of therapeutics, Clin. Pharmacol. Ther, doi:10.1002/cpt.2160
Perelson, Ribeiro, Phan, An explanation for SARS-CoV-2 rebound after Paxlovid treatment, medRxiv, doi:10.1101/2023.05.30.23290747
Phan, Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody, bioRxiv, doi:10.1101/2023.09.14.557679
Ranard, Chow, Megjhani, Asgari, Park et al., A mathematical model of SARS-CoV-2 immunity predicts paxlovid rebound, J. Med. Virol, doi:10.1002/jmv.28854
Rhee, Kanjilal, Baker, Klompas, Duration of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation?, Clin. Infect. Dis, doi:10.1093/cid/ciaa1249
Ruysseveldt, Martens, Steelant, Airway basal cells, protectors of epithelial walls in health and respiratory diseases, Front. Allergy, doi:10.3389/falgy.2021.787128
Schuh, Markov, Veliov, Stilianakis, A mathematical model for the within-host (re)infection dynamics of SARS-CoV-2, Math. Biosci, doi:10.1016/j.mbs.2024.109178
Schuh, Markov, Voulgaridi, Bogogiannidou, Mouchtouri et al., Supplementary material from: Within-host dynamics of antiviral treatment with remdesivir for SARS-CoV-2 infection, Figshare, doi:10.1101/2024.05.31.24308284
Spinner, Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19, JAMA, doi:10.1001/jama.2020.16349
Svanberg, Early stimulated immune responses predict clinical disease severity in hospitalized COVID-19 patients, Commun. Med, doi:10.1038/s43856-022-00178-5
Szablewski, SARS-CoV-2 transmission and infection among attendees of an overnight camp -Georgia, June 2020, MMWR Morb. Mortal. Wkly. Rep, doi:10.15585/mmwr.mm6931e1
Tanni, Silvinato, Floriano, Bacha, Barbosa et al., Use of remdesivir in patients with COVID-19: a systematic review and meta-analysis, J. Bras. Pneumol, doi:10.36416/1806-3756/e20210393
Wang, Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial, The Lancet, doi:10.1016/S0140-6736(20)31022-9
Wu, Kang, Guo, Liu, Liu et al., Incubation period of COVID-19 caused by unique SARS-CoV-2 strains: a systematic review and meta-analysis, JAMA Netw. Open, doi:10.1001/jamanetworkopen.2022.28008
Wölfel, Virological assessment of hospitalized patients with COVID-2019, Nature, doi:10.1038/s41586-020-2196-x
Zitzmann, Ke, Ribeiro, Perelson, How robust are estimates of key parameters in standard viral dynamic models?, PLoS Comput. Biol, doi:10.1371/journal.pcbi.1011437
DOI record:
{
"DOI": "10.1098/rsif.2024.0536",
"ISSN": [
"1742-5689",
"1742-5662"
],
"URL": "http://dx.doi.org/10.1098/rsif.2024.0536",
"abstract": "<jats:p>The effectiveness of antiviral treatment with remdesivir against COVID-19 has been investigated in clinical trials suggesting earlier recovery. However, this effect seems to be rather modest. In this study, we tracked the clinical course of SARS-CoV-2 infections in 369 COVID-19 individuals across a spectrum of illness severities, including both untreated individuals and individuals who received antiviral treatment with remdesivir. Moreover, using a process-based mathematical model, we quantified and analysed the within-host infection dynamics of a total of 88 individuals, of which 69 were untreated and 19 antiviral-treated individuals. For untreated individuals, we found that those hospitalized exhibit lower levels of early immune response and higher cumulative viral loads than those who were not. For treated individuals, we found that those who died were on average hospitalized later after symptom onset than those who survived, underscoring the importance of early medical intervention for severe COVID-19. Finally, our model estimates a rather limited antiviral activity of remdesivir. Our results provide valuable insights into the clinical course of COVID-19 during antiviral treatment with remdesivir and suggest the need for alternative treatment regimens.</jats:p>",
"alternative-id": [
"10.1098/rsif.2024.0536"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-08-06"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2024-10-09"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2024-10-28"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2024-11-27"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-3721-7532",
"affiliation": [
{
"name": "Joint Research Centre (JRC), European Commission, Ispra, Italy"
}
],
"authenticated-orcid": true,
"family": "Schuh",
"given": "Lea",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Joint Research Centre (JRC), European Commission, Ispra, Italy"
},
{
"name": "London School of Hygiene & Tropical Medicine, University of London, London, UK"
}
],
"family": "Markov",
"given": "Peter V.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Hygiene and Epidemiology, University of Thessaly, Larissa, Greece"
}
],
"family": "Voulgaridi",
"given": "Ioanna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Hygiene and Epidemiology, University of Thessaly, Larissa, Greece"
}
],
"family": "Bogogiannidou",
"given": "Zacharoula",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Hygiene and Epidemiology, University of Thessaly, Larissa, Greece"
}
],
"family": "Mouchtouri",
"given": "Varvara A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of Hygiene and Epidemiology, University of Thessaly, Larissa, Greece"
}
],
"family": "Hadjichristodoulou",
"given": "Christos",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3808-265X",
"affiliation": [
{
"name": "Joint Research Centre (JRC), European Commission, Ispra, Italy"
},
{
"name": "Department of Biometry and Epidemiology, University of Erlangen-Nuremberg, Erlangen, Germany"
}
],
"authenticated-orcid": true,
"family": "Stilianakis",
"given": "Nikolaos I.",
"sequence": "additional"
}
],
"container-title": "Journal of The Royal Society Interface",
"container-title-short": "J. R. Soc. Interface.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"royalsocietypublishing.org"
]
},
"created": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T00:05:13Z",
"timestamp": 1732665913000
},
"deposited": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T00:05:23Z",
"timestamp": 1732665923000
},
"indexed": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T05:38:20Z",
"timestamp": 1732685900486,
"version": "3.28.2"
},
"is-referenced-by-count": 0,
"issue": "220",
"issued": {
"date-parts": [
[
2024,
11
]
]
},
"journal-issue": {
"issue": "220",
"published-print": {
"date-parts": [
[
2024,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
1
]
],
"date-time": "2024-11-01T00:00:00Z",
"timestamp": 1730419200000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
1
]
],
"date-time": "2024-11-01T00:00:00Z",
"timestamp": 1730419200000
}
}
],
"link": [
{
"URL": "https://royalsocietypublishing.org/doi/pdf/10.1098/rsif.2024.0536",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://royalsocietypublishing.org/doi/full-xml/10.1098/rsif.2024.0536",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://royalsocietypublishing.org/doi/pdf/10.1098/rsif.2024.0536",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "175",
"original-title": [],
"prefix": "10.1098",
"published": {
"date-parts": [
[
2024,
11
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
27
]
]
},
"published-print": {
"date-parts": [
[
2024,
11
]
]
},
"publisher": "The Royal Society",
"reference": [
{
"DOI": "10.1056/NEJMoa2007764",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_2_2"
},
{
"DOI": "10.36416/1806-3756/e20210393",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_3_2"
},
{
"DOI": "10.1001/jama.2020.16349",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_4_2"
},
{
"DOI": "10.1016/S0140-6736(20)31022-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_5_2"
},
{
"DOI": "10.1056/NEJMoa2023184",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_6_2"
},
{
"DOI": "10.1016/S1473-3099(22)00295-X",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_7_2"
},
{
"DOI": "10.7326/M21-0653",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_8_2"
},
{
"DOI": "10.1038/s41586-020-2196-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_9_2"
},
{
"DOI": "10.1073/pnas.2017962118",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_10_2"
},
{
"DOI": "10.1172/jci.insight.176286",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_11_2"
},
{
"DOI": "10.1002/jmv.27598",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_12_2"
},
{
"DOI": "10.3390/microorganisms11020312",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_13_2"
},
{
"DOI": "10.1016/j.cmi.2021.02.029",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_14_2"
},
{
"article-title": "Modeling explains prolonged SARS-CoV-2 nasal shedding relative to lung shedding in remdesivir-treated rhesus macaques",
"author": "Goyal A",
"journal-title": "i. Sci.",
"key": "e_1_3_8_15_2",
"unstructured": "Goyal A, Duke ER, Cardozo-Ojeda EF, Schiffer JT. 2022 Modeling explains prolonged SARS-CoV-2 nasal shedding relative to lung shedding in remdesivir-treated rhesus macaques. i. Sci. 25, 104448. (doi:10.1016/j.isci.2022.104448)",
"volume": "25",
"year": "2022"
},
{
"DOI": "10.1186/s12916-021-02220-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_16_2"
},
{
"DOI": "10.1038/s41564-022-01105-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_17_2"
},
{
"DOI": "10.1002/cpt.2160",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_18_2"
},
{
"DOI": "10.1016/j.mbs.2024.109178",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_19_2"
},
{
"DOI": "10.1093/jac/dkac048",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_20_2"
},
{
"article-title": "Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody",
"author": "Phan T",
"journal-title": "bioRxiv.",
"key": "e_1_3_8_21_2",
"unstructured": "Phan T et al. 2023 Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody. bioRxiv. (doi:10.1101/2023.09.14.557679)",
"year": "2023"
},
{
"DOI": "10.3389/falgy.2021.787128",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_22_2"
},
{
"DOI": "10.15585/mmwr.mm6931e1",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_23_2"
},
{
"DOI": "10.2139/ssrn.3716879",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_24_2"
},
{
"DOI": "10.1016/j.ijid.2021.01.069",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_25_2"
},
{
"DOI": "10.1371/journal.pcbi.1011437",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_26_2"
},
{
"DOI": "10.1093/cid/ciaa1249",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_27_2"
},
{
"author": "Centers for Disease Control and Prevention",
"key": "e_1_3_8_28_2",
"unstructured": "Centers for Disease Control and Prevention. 2023 Ending isolation and precautions for people with COVID-19: interim guidance. See https://archive.cdc.gov/#/details?q=https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html&start=0&rows=10&url=https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html (accessed 14 November 2024).",
"volume-title": "Ending isolation and precautions for people with COVID-19: interim guidance",
"year": "2023"
},
{
"DOI": "10.1038/s43856-022-00178-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_29_2"
},
{
"article-title": "Nonlinear mixed models and related approaches in infectious disease modeling: a systematic and critical review",
"author": "Adéoti OM",
"first-page": "110",
"journal-title": "Infect. Dis. Model.",
"key": "e_1_3_8_30_2",
"unstructured": "Adéoti OM, Agbla S, Diop A, Glèlè Kakaï R. 2025 Nonlinear mixed models and related approaches in infectious disease modeling: a systematic and critical review. Infect. Dis. Model. 10, 110–128. (doi:10.1016/j.idm.2024.09.001)",
"volume": "10",
"year": "2025"
},
{
"DOI": "10.1001/jamanetworkopen.2022.28008",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_31_2"
},
{
"DOI": "10.1073/pnas.2111477118",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_32_2"
},
{
"DOI": "10.1371/journal.pcbi.1010721",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_33_2"
},
{
"DOI": "10.1002/jmv.28854",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_34_2"
},
{
"article-title": "An explanation for SARS-CoV-2 rebound after Paxlovid treatment",
"author": "Perelson AS",
"journal-title": "medRxiv",
"key": "e_1_3_8_35_2",
"unstructured": "Perelson AS, Ribeiro RM, Phan T. 2023 An explanation for SARS-CoV-2 rebound after Paxlovid treatment. medRxiv 2023.05.30.23290747. (doi:10.1101/2023.05.30.23290747)",
"year": "2023"
},
{
"key": "e_1_3_8_36_2",
"unstructured": "The MathWorks Inc. binornd: random numbers from binomial distribution. See https://it.mathworks.com/help/stats/binornd.html."
},
{
"key": "e_1_3_8_37_2",
"unstructured": "cdf. 2024 Cumulative distribution function - MATLAB - MathWorks Italia. See https://it.mathworks.com/help/stats/prob.normaldistribution.cdf.html (accessed 27 September 2024)."
},
{
"DOI": "10.1038/s41467-020-20542-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_38_2"
},
{
"DOI": "10.1093/bioinformatics/btz020",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_39_2"
},
{
"key": "e_1_3_8_40_2",
"unstructured": "The MathWorks Inc. fmincon - find minimum of constrained nonlinear multivariable function. See https://it.mathworks.com/help/optim/ug/fmincon.html."
},
{
"key": "e_1_3_8_41_2",
"unstructured": "The MathWorks Inc. ode45 - solve nonstiff differential equations — medium order method. See https://it.mathworks.com/help/matlab/ref/ode45.html."
},
{
"DOI": "10.1080/01621459.1995.10476572",
"doi-asserted-by": "publisher",
"key": "e_1_3_8_42_2"
},
{
"key": "e_1_3_8_43_2",
"unstructured": "MathWorks. kstest2 - Two-sample Kolmogorov-Smirnov test. See https://it.mathworks.com/help/stats/kstest2.html."
},
{
"key": "e_1_3_8_44_2",
"unstructured": "Keine C. Mood’s median test. See https://it.mathworks.com/matlabcentral/fileexchange/70081-moods-median-test."
},
{
"key": "e_1_3_8_45_2",
"unstructured": "lea-schuh. 2024 lea-schuh/COVID_treat: v1.0.0 (v1.0.0). Zenodo (doi:10.5281/zenodo.13905096)"
},
{
"DOI": "10.1101/2024.05.31.24308284",
"doi-asserted-by": "crossref",
"key": "e_1_3_8_46_2",
"unstructured": "Schuh L Markov PV Voulgaridi I Bogogiannidou Z Mouchtouri VA Hadjichristodoulou C et al. 2024 Supplementary material from: Within-host dynamics of antiviral treatment with remdesivir for SARS-CoV-2 infection. Figshare. (doi:10.1101/2024.05.31.24308284)"
}
],
"reference-count": 45,
"references-count": 45,
"relation": {},
"resource": {
"primary": {
"URL": "https://royalsocietypublishing.org/doi/10.1098/rsif.2024.0536"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Within-host dynamics of antiviral treatment with remdesivir for SARS-CoV-2 infection",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1098/crossmark-policy",
"volume": "21"
}