In Silico Evaluation of Structural Consequences in the Human CYP3A4 Caused by Molnupiravir-Induced Mutations During COVID-19 Treatment
et al., Drugs and Drug Candidates, doi:10.3390/ddc4040050, Nov 2025
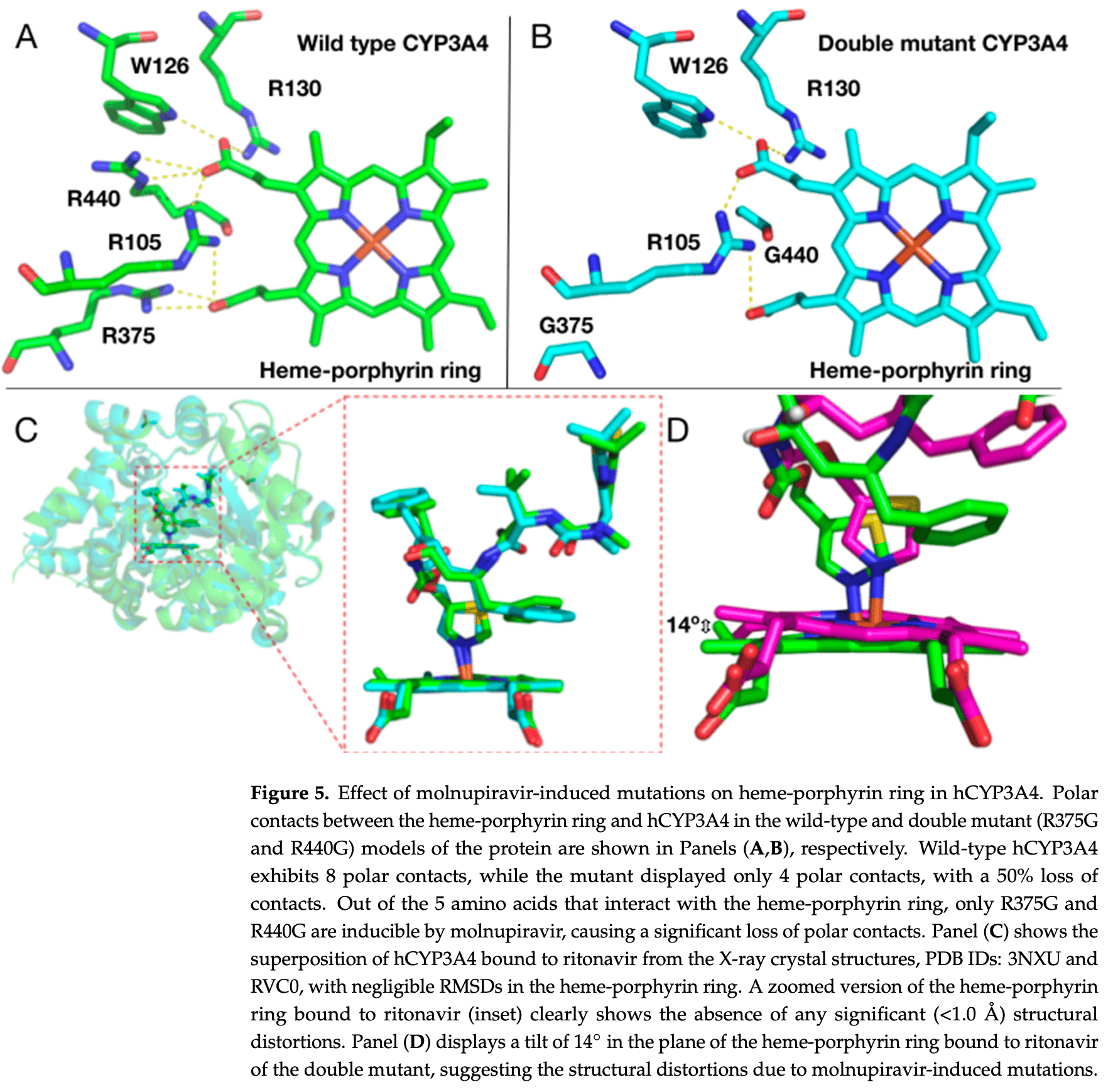
In silico study showing potential harm from molnupiravir-induced mutations in human CYP3A4 enzyme during COVID-19 treatment. Authors identified six hotspot amino acids (R105, W126, R130, R375, S437, R440) in the CYP3A4 active site that could be mutated by molnupiravir, with mutations at positions 105, 126, and 130 causing premature stop codons resulting in severely truncated, non-functional enzyme variants. The study suggests that molnupiravir-induced mutations could impair CYP3A4 function, potentially leading to inefficient drug metabolism and accumulation of medications in COVID-19 patients with comorbidities, particularly affecting the efficacy of paxlovid treatment where ritonavir relies on CYP3A4 inhibition to boost nirmatrelvir levels.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Aggunna et al., 11 Nov 2025, India, peer-reviewed, 11 authors.
Contact: tcabse.india@gmail.com (corresponding author), aggunnamadhumita@gmail.com, gantetichiranjeevi42@gmail.com, keerthirathod91@gmail.com, minnimeghana2@gmail.com, joyjethinneelam@gmail.com, aswithagurrala@gmail.com, grandhibavana@gmail.com, noahjeevanvejendla@gmail.com, kruparabbuni@gmail.com.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
In Silico Evaluation of Structural Consequences in the Human CYP3A4 Caused by Molnupiravir-Induced Mutations During COVID-19 Treatment
Drugs and Drug Candidates, doi:10.3390/ddc4040050
Background/Objectives: Molnupiravir (MOV) and nirmatrelvir (NMV) are antiviral drugs that were FDA-approved under the emergency use authorization (EUA) for coronavirus disease-2019 (COVID-19) treatment. MOV and NMV target the viral RNA-dependent RNA polymerase and main protease, respectively. Paxlovid is a combination of NMV and ritonavir (RTV), an inhibitor of the human cytochrome P450-3A4 (hCYP3A4). In this study, the structural consequences in the hCYP3A4 caused by MOV-induced mutations (MIM) were evaluated using in silico tools. Methods: MOV-induced mutations (MIM) were inserted into all the possible hotspots in the active site region of the hCYP3A4 gene, and mutant protein models were built. Structural changes in the heme-porphyrin ring of hCYP3A4 were analyzed in the presence and absence of substrates/inhibitors, including RTV. Molecular dynamics (MD) simulations were performed to analyze the effect of MIMinduced structural changes in hCYP3A4 on drug binding. Results: MD simulations confirm that MIMs, R375G and R440G in hCYP3A4 severely affect the heme-porphyrin ring stability by causing a tilt that in turn affects RTV binding, suggesting a possible inefficiency in the function of hCYP3A4. Similar results were seen for amlodipine, atorvastatin, sildenafil and warfarin, which are substrates of hCYP3A4. Conclusions: The current in silico studies indicate that hCYP3A4 containing MIMs can create complications in the treatment of COVID-19 patients, particularly with co-morbidities due to its functional inefficiency. Hence, clinicians must be vigilant when using MOV in combination with other drugs. Further in vitro studies focused on hCYP3A4 containing MIMs are currently in progress to support our current in silico findings.
Author Contributions: M.A., A.G., B.G., J.N., M.M., N.V. and S.M. generated all the mutant models of hCYP3A4 with MIMs, analyzed them by MD simulations and plotted the trajectory data for hCYP3A4; C.V.M.G. and K.R.B. performed docking and MD simulations of hCYP3A4 ligands; S.G. co-supervised A.G., B.G., J.N., M.M., N.V. and S.M.; R.S.Y. is the principal investigator who designed the study and supervised M.A., C.V.M.G., K.R.B., A.G., B.G., J.N., M.M., N.V. and S.M.; R.S.Y. is responsible for project administration, funding acquisition, conceptualization, writing, editing and finalizing the manuscript. All authors have read and agreed to the published version of the manuscript. Funding: This research received no external funding.
Conflicts of Interest: The authors in this manuscript declare no conflicts of interest.
References
Aboul-Fotouh, Mahmoud, Elnahas, Habib, Abdelraouf, What are the current anti-COVID-19 drugs? From traditional to smart molecular mechanisms, Virol. J, doi:10.1186/s12985-023-02210-z
Addala, Vissapragada, Aggunna, Mukala, Lanka et al., Success of Current COVID-19 Vaccine Strategies vs. the Epitope Topology of SARS-CoV-2 Spike Protein-Receptor Binding Domain (RBD): A Computational Study of RBD Topology to Guide Future Vaccine Design, Vaccines, doi:10.3390/vaccines10060841
Aggunna, Grandhi, Yedidi, Molecular dynamics simulations of cytotoxin-associated gene A coded protein from Helicobacter pylori to probe the flexibility of p53 binding pocket for inhibitor design, TCABSE-J
Ashour, Elmaaty, Sarhan, Elkaeed, Moussa et al., A Systematic Review of the Global Intervention for SARS-CoV-2 Combating: From Drugs Repurposing to Molnupiravir Approval, Drug Des. Dev. Ther, doi:10.2147/DDDT.S354841
Bhardwaj, Kour, Rai, Bhattacharya, Manhas et al., Treatment Tweaks CYP3A4 and CYP2C8 in Arthritic Rats to Expedite Drug Interaction: Implication in Oral Therapy of Molnupiravir, ACS Omega, doi:10.1021/acsomega.3c09287
Carbone, Paradis, Brunt, Wu, Binding Mechanism of the Active Form of Molnupiravir to RdRp of SARS-CoV-2 and Designing Potential Analogues: Insights from Molecular Dynamics Simulations, ACS Omega, doi:10.1021/acsomega.4c05469
Charness, Gupta, Stack, Strymish, Adams et al., Rebound of SARS-CoV-2 Infection after Nirmatrelvir-Ritonavir Treatment, N. Engl. J. Med, doi:10.1056/NEJMc2206449
Elshaboury, Monk, Bebell, Bidell, Adamsick et al., Remdesivir use and outcomes during the FDA COVID-19 emergency use authorization period, Ther. Adv. Infect. Dis, doi:10.1177/20499361211046669
Esmaesmaeili, Owens, Wagoner, Polyak, White et al., A unifying model to explain frequent SARS-CoV-2 rebound after nirmatrelvir treatment and limited prophylactic efficacy, Nat. Commun, doi:10.1038/s41467-024-49458-9
Gasteiger, Gattiker, Hoogland, Ivanyi, Appel et al., ExPASy: The proteomics server for in-depth protein knowledge and analysis, Nucleic Acids Res, doi:10.1093/nar/gkg563
Ghosh, Mishevich, Mesecar, Mitsuya, Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19, ChemMedChem, doi:10.1002/cmdc.202200440
Goodman, Woodgate, Translesion DNA polymerases, Cold Spring Harb. Perspect. Biol, doi:10.1101/cshperspect.a010363
Gordon, Tchesnokov, Woolner, Perry, Feng et al., Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency, J. Biol. Chem, doi:10.1074/jbc.RA120.013679
Hammond, Yunis, Fountaine, Luscan, Burr et al., Oral Nirmatrelvir-Ritonavir as Postexposure Prophylaxis for Covid-19, N. Engl. J. Med, doi:10.1056/NEJMoa2309002
Harris, FDA Grants Full Approval to Paxlovid, COVID-19 Antiviral Treatment, JAMA, doi:10.1001/jama.2023.9925
Holman, Holman, Mcintosh, Painter, Painter et al., Accelerated first-in-human clinical trial of EIDD-2801/MK-4482 (molnupiravir), a ribonucleoside analog with potent antiviral activity against SARS-CoV-2, Trials, doi:10.1186/s13063-021-05538-5
Hsu, Savas, Johnson, The X-Ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5, Mol. Pharmacol, doi:10.1124/mol.117.109744
Kabinger, Stiller, Schmitzová, Dienemann, Kokic et al., Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00651-0
Kim, Yoon, Lee, Investigating the Safety Profile of Fast-Track COVID-19 Drugs Using the FDA Adverse Event Reporting System Database: A Comparative Observational Study, Pharmacoepidemiol. Drug Saf, doi:10.1002/pds.70043
Lackey, Thompson, Eggers, FDA's Benefit-Risk Framework for Human Drugs and Biologics: Role in Benefit-Risk Assessment and Analysis of Use for Drug Approvals, Ther. Innov. Regul. Sci, doi:10.1007/s43441-020-00203-6
Lobinska, Pilpel, Nowak, Evolutionary safety of lethal mutagenesis driven by antiviral treatment, PLoS Biol, doi:10.1371/journal.pbio.3002214
Purandare, Understanding Drug Development: A Primer on the Food and Drug Administration, J. Pediatr. Infect. Dis. Soc, doi:10.1093/jpids/piab023
Rizk, Forthal, Kalantar-Zadeh, Mehra, Lavie et al., Expanded Access Programs, compassionate drug use, and Emergency Use Authorizations during the COVID-19 pandemic, Drug Discov. Today, doi:10.1016/j.drudis.2020.11.025
Samuels, Sevrioukova, An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4, Bioorganic Med. Chem, doi:10.1016/j.bmc.2020.115349
Sanderson, Hisner, Donovan-Banfield, Hartman, Løchen et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6
Schaefer, Cheng, Recent Advances in Covalent Drug Discovery, Pharmaceuticals, doi:10.3390/ph16050663
Sevrioukova, High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4, Biochemistry, doi:10.1021/acs.biochem.7b00334
Sevrioukova, Poulos, Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1010693107
Tan, Joyce, Tan, Hu, Wang, SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies, Accounts Chem. Res, doi:10.1021/acs.accounts.2c00735
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem, doi:10.1002/jcc.21334
Vankadara, Dawson, Fong, Oh, Ang et al., A Warhead Substitution Study on the Coronavirus Main Protease Inhibitor Nirmatrelvir, ACS Med. Chem. Lett, doi:10.1021/acsmedchemlett.2c00260
Vissapragada, Addala, Aggunna, Mukala, Chintalapati et al., In silico analysis of molnupiravir usage vs. efficacy of COVID-19 vaccines, TCABSE-J
Vissapragada, Aggunna, Tallapalli, Mandugula, Devandla et al., In silico prediction of COVID-19 vaccine efficacy based on the strain-specific structural deviations in the SARS CoV-2 spike protein receptor binding domain, Med. Res. Arch, doi:10.18103/mra.v12i9.5718
Waterhouse, Bertoni, Bienert, Studer, Tauriello et al., SWISS-MODEL: Homology modelling of protein structures and complexes, Nucleic Acids Res, doi:10.1093/nar/gky427
Waters, Warren, A tale of two drugs: Molnupiravir and Paxlovid, Mutat. Res. Mol. Mech. Mutagen, doi:10.1016/j.mrrev.2025.108533
DOI record:
{
"DOI": "10.3390/ddc4040050",
"ISSN": [
"2813-2998"
],
"URL": "http://dx.doi.org/10.3390/ddc4040050",
"abstract": "<jats:p>Background/Objectives: Molnupiravir (MOV) and nirmatrelvir (NMV) are antiviral drugs that were FDA-approved under the emergency use authorization (EUA) for coronavirus disease-2019 (COVID-19) treatment. MOV and NMV target the viral RNA-dependent RNA polymerase and main protease, respectively. Paxlovid is a combination of NMV and ritonavir (RTV), an inhibitor of the human cytochrome P450-3A4 (hCYP3A4). In this study, the structural consequences in the hCYP3A4 caused by MOV-induced mutations (MIM) were evaluated using in silico tools. Methods: MOV-induced mutations (MIM) were inserted into all the possible hotspots in the active site region of the hCYP3A4 gene, and mutant protein models were built. Structural changes in the heme-porphyrin ring of hCYP3A4 were analyzed in the presence and absence of substrates/inhibitors, including RTV. Molecular dynamics (MD) simulations were performed to analyze the effect of MIM-induced structural changes in hCYP3A4 on drug binding. Results: MD simulations confirm that MIMs, R375G and R440G in hCYP3A4 severely affect the heme-porphyrin ring stability by causing a tilt that in turn affects RTV binding, suggesting a possible inefficiency in the function of hCYP3A4. Similar results were seen for amlodipine, atorvastatin, sildenafil and warfarin, which are substrates of hCYP3A4. Conclusions: The current in silico studies indicate that hCYP3A4 containing MIMs can create complications in the treatment of COVID-19 patients, particularly with co-morbidities due to its functional inefficiency. Hence, clinicians must be vigilant when using MOV in combination with other drugs. Further in vitro studies focused on hCYP3A4 containing MIMs are currently in progress to support our current in silico findings.</jats:p>",
"alternative-id": [
"ddc4040050"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-0174-2965",
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "School of Medical Sciences, Örebro University, 70382 Örebro, Sweden"
}
],
"authenticated-orcid": false,
"family": "Aggunna",
"given": "Madhumita",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
}
],
"family": "Ganteti",
"given": "Chiranjeevi V. M.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
}
],
"family": "Bhukya",
"given": "Keerthi R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Mathangi",
"given": "Meghana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Neelam",
"given": "Joyjethin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Gurrala",
"given": "Aswitha",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Grandhi",
"given": "Bavana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Vejendla",
"given": "Noahjeevan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Mathangi",
"given": "Sriharshini",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Biomedical Sciences, Hindu College, Guntur 522002, India"
}
],
"family": "Gudapati",
"given": "Swarnalatha",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2755-1307",
"affiliation": [
{
"name": "Department of Intramural Research Core, The Center for Advanced-Applied Biological Sciences & Entrepreneurship (TCABS-E), Visakhapatnam 530003, India"
},
{
"name": "Department of Biotechnology, Andhra University, Visakhapatnam 530003, India"
}
],
"authenticated-orcid": false,
"family": "Yedidi",
"given": "Ravikiran S.",
"sequence": "additional"
}
],
"container-title": "Drugs and Drug Candidates",
"container-title-short": "DDC",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
11,
11
]
],
"date-time": "2025-11-11T10:56:27Z",
"timestamp": 1762858587000
},
"deposited": {
"date-parts": [
[
2025,
11,
11
]
],
"date-time": "2025-11-11T11:36:14Z",
"timestamp": 1762860974000
},
"indexed": {
"date-parts": [
[
2025,
11,
11
]
],
"date-time": "2025-11-11T11:39:27Z",
"timestamp": 1762861167238,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "4",
"issued": {
"date-parts": [
[
2025,
11,
11
]
]
},
"journal-issue": {
"issue": "4",
"published-online": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
11,
11
]
],
"date-time": "2025-11-11T00:00:00Z",
"timestamp": 1762819200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2813-2998/4/4/50/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "50",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
11,
11
]
]
},
"published-online": {
"date-parts": [
[
2025,
11,
11
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1007/s43441-020-00203-6",
"article-title": "FDA’s Benefit–Risk Framework for Human Drugs and Biologics: Role in Benefit–Risk Assessment and Analysis of Use for Drug Approvals",
"author": "Lackey",
"doi-asserted-by": "crossref",
"first-page": "170",
"journal-title": "Ther. Innov. Regul. Sci.",
"key": "ref_1",
"volume": "55",
"year": "2021"
},
{
"DOI": "10.1093/jpids/piab023",
"article-title": "Understanding Drug Development: A Primer on the Food and Drug Administration",
"author": "Purandare",
"doi-asserted-by": "crossref",
"first-page": "977",
"journal-title": "J. Pediatr. Infect. Dis. Soc.",
"key": "ref_2",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1016/j.drudis.2020.11.025",
"article-title": "Expanded Access Programs, compassionate drug use, and Emergency Use Authorizations during the COVID-19 pandemic",
"author": "Rizk",
"doi-asserted-by": "crossref",
"first-page": "593",
"journal-title": "Drug Discov. Today",
"key": "ref_3",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1002/pds.70043",
"article-title": "Investigating the Safety Profile of Fast-Track COVID-19 Drugs Using the FDA Adverse Event Reporting System Database: A Comparative Observational Study",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "e70043",
"journal-title": "Pharmacoepidemiol. Drug Saf.",
"key": "ref_4",
"volume": "33",
"year": "2024"
},
{
"DOI": "10.1186/s12985-023-02210-z",
"article-title": "What are the current anti-COVID-19 drugs? From traditional to smart molecular mechanisms",
"author": "Mahmoud",
"doi-asserted-by": "crossref",
"first-page": "241",
"journal-title": "Virol. J.",
"key": "ref_5",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.1002/cmdc.202200440",
"article-title": "Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19",
"author": "Ghosh",
"doi-asserted-by": "crossref",
"first-page": "e202200440",
"journal-title": "ChemMedChem",
"key": "ref_6",
"volume": "17",
"year": "2022"
},
{
"article-title": "Remdesivir use and outcomes during the FDA COVID-19 emergency use authorization period",
"author": "Elshaboury",
"first-page": "20499361211046669",
"journal-title": "Ther. Adv. Infect. Dis.",
"key": "ref_7",
"volume": "8",
"year": "2021"
},
{
"DOI": "10.1074/jbc.RA120.013679",
"article-title": "Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency",
"author": "Gordon",
"doi-asserted-by": "crossref",
"first-page": "6785",
"journal-title": "J. Biol. Chem.",
"key": "ref_8",
"volume": "295",
"year": "2020"
},
{
"DOI": "10.1021/acsomega.4c05469",
"article-title": "Binding Mechanism of the Active Form of Molnupiravir to RdRp of SARS-CoV-2 and Designing Potential Analogues: Insights from Molecular Dynamics Simulations",
"author": "Carbone",
"doi-asserted-by": "crossref",
"first-page": "41583",
"journal-title": "ACS Omega",
"key": "ref_9",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.1186/s13063-021-05538-5",
"article-title": "Accelerated first-in-human clinical trial of EIDD-2801/MK-4482 (molnupiravir), a ribonucleoside analog with potent antiviral activity against SARS-CoV-2",
"author": "Holman",
"doi-asserted-by": "crossref",
"first-page": "561",
"journal-title": "Trials",
"key": "ref_10",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1021/acsmedchemlett.2c00260",
"article-title": "A Warhead Substitution Study on the Coronavirus Main Protease Inhibitor Nirmatrelvir",
"author": "Vankadara",
"doi-asserted-by": "crossref",
"first-page": "1345",
"journal-title": "ACS Med. Chem. Lett.",
"key": "ref_11",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.3390/ph16050663",
"doi-asserted-by": "crossref",
"key": "ref_12",
"unstructured": "Schaefer, D., and Cheng, X. (2023). Recent Advances in Covalent Drug Discovery. Pharmaceuticals, 16."
},
{
"article-title": "A tale of two drugs: Molnupiravir and Paxlovid",
"author": "Waters",
"first-page": "108533",
"journal-title": "Mutat. Res. Mol. Mech. Mutagen.",
"key": "ref_13",
"volume": "795",
"year": "2025"
},
{
"DOI": "10.1021/acs.accounts.2c00735",
"article-title": "SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies",
"author": "Tan",
"doi-asserted-by": "crossref",
"first-page": "157",
"journal-title": "Accounts Chem. Res.",
"key": "ref_14",
"volume": "56",
"year": "2023"
},
{
"DOI": "10.2147/DDDT.S354841",
"article-title": "A Systematic Review of the Global Intervention for SARS-CoV-2 Combating: From Drugs Repurposing to Molnupiravir Approval",
"author": "Ashour",
"doi-asserted-by": "crossref",
"first-page": "685",
"journal-title": "Drug Des. Dev. Ther.",
"key": "ref_15",
"volume": "16",
"year": "2022"
},
{
"DOI": "10.1073/pnas.1010693107",
"article-title": "Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir",
"author": "Sevrioukova",
"doi-asserted-by": "crossref",
"first-page": "18422",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_16",
"volume": "107",
"year": "2010"
},
{
"DOI": "10.1124/mol.117.109744",
"article-title": "The X-Ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5",
"author": "Hsu",
"doi-asserted-by": "crossref",
"first-page": "14",
"journal-title": "Mol. Pharmacol.",
"key": "ref_17",
"volume": "93",
"year": "2018"
},
{
"article-title": "FDA Grants Full Approval to Paxlovid, COVID-19 Antiviral Treatment",
"author": "Harris",
"first-page": "2118",
"journal-title": "JAMA",
"key": "ref_18",
"volume": "329",
"year": "2023"
},
{
"DOI": "10.1038/s41594-021-00651-0",
"article-title": "Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis",
"author": "Kabinger",
"doi-asserted-by": "crossref",
"first-page": "740",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_19",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1371/journal.pbio.3002214",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Lobinska, G., Pilpel, Y., and Nowak, M.A. (2023). Evolutionary safety of lethal mutagenesis driven by antiviral treatment. PLoS Biol., 21."
},
{
"DOI": "10.1056/NEJMoa2309002",
"article-title": "Oral Nirmatrelvir–Ritonavir as Postexposure Prophylaxis for Covid-19",
"author": "Hammond",
"doi-asserted-by": "crossref",
"first-page": "224",
"journal-title": "N. Engl. J. Med.",
"key": "ref_21",
"volume": "391",
"year": "2024"
},
{
"DOI": "10.1038/s41467-024-49458-9",
"article-title": "A unifying model to explain frequent SARS-CoV-2 rebound after nirmatrelvir treatment and limited prophylactic efficacy",
"author": "EsmaEsmaeili",
"doi-asserted-by": "crossref",
"first-page": "5478",
"journal-title": "Nat. Commun.",
"key": "ref_22",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1021/acsomega.3c09287",
"article-title": "EIDD-1931 Treatment Tweaks CYP3A4 and CYP2C8 in Arthritic Rats to Expedite Drug Interaction: Implication in Oral Therapy of Molnupiravir",
"author": "Bhardwaj",
"doi-asserted-by": "crossref",
"first-page": "13982",
"journal-title": "ACS Omega",
"key": "ref_23",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.1038/s41586-023-06649-6",
"article-title": "A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes",
"author": "Sanderson",
"doi-asserted-by": "crossref",
"first-page": "594",
"journal-title": "Nature",
"key": "ref_24",
"volume": "623",
"year": "2023"
},
{
"article-title": "In silico analysis of molnupiravir usage vs. efficacy of COVID-19 vaccines",
"author": "Vissapragada",
"first-page": "1",
"journal-title": "TCABSE-J.",
"key": "ref_25",
"volume": "1",
"year": "2023"
},
{
"DOI": "10.3390/vaccines10060841",
"doi-asserted-by": "crossref",
"key": "ref_26",
"unstructured": "Addala, S., Vissapragada, M., Aggunna, M., Mukala, N., Lanka, M., Gampa, S., Sodasani, M., Chintalapati, J., Kamidi, A., and Veeranna, R.P. (2022). Success of Current COVID-19 Vaccine Strategies vs. the Epitope Topology of SARS-CoV-2 Spike Protein-Receptor Binding Domain (RBD): A Computational Study of RBD Topology to Guide Future Vaccine Design. Vaccines, 10."
},
{
"DOI": "10.18103/mra.v12i9.5718",
"doi-asserted-by": "crossref",
"key": "ref_27",
"unstructured": "Vissapragada, M., Aggunna, M., Tallapalli, M., Mandugula, H., Devandla, A., Yekula, A., Malapati, A., Bonala, S., Addala, S., and Gudapati, S. (2024). In silico prediction of COVID-19 vaccine efficacy based on the strain-specific structural deviations in the SARS CoV-2 spike protein receptor binding domain. Med. Res. Arch., 12."
},
{
"DOI": "10.1056/NEJMc2206449",
"article-title": "Rebound of SARS-CoV-2 Infection after Nirmatrelvir–Ritonavir Treatment",
"author": "Charness",
"doi-asserted-by": "crossref",
"first-page": "1045",
"journal-title": "N. Engl. J. Med.",
"key": "ref_28",
"volume": "387",
"year": "2022"
},
{
"DOI": "10.1101/cshperspect.a010363",
"doi-asserted-by": "crossref",
"key": "ref_29",
"unstructured": "Goodman, M.F., and Woodgate, R. (2013). Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol., 5."
},
{
"DOI": "10.1093/nar/gkg563",
"article-title": "ExPASy: The proteomics server for in-depth protein knowledge and analysis",
"author": "Gasteiger",
"doi-asserted-by": "crossref",
"first-page": "3784",
"journal-title": "Nucleic Acids Res.",
"key": "ref_30",
"volume": "31",
"year": "2003"
},
{
"DOI": "10.1093/nar/gky427",
"article-title": "SWISS-MODEL: Homology modelling of protein structures and complexes",
"author": "Waterhouse",
"doi-asserted-by": "crossref",
"first-page": "W296",
"journal-title": "Nucleic Acids Res.",
"key": "ref_31",
"volume": "46",
"year": "2018"
},
{
"article-title": "Molecular dynamics simulations of cytotoxin-associated gene A coded protein from Helicobacter pylori to probe the flexibility of p53 binding pocket for inhibitor design",
"author": "Aggunna",
"first-page": "9",
"journal-title": "TCABSE-J.",
"key": "ref_32",
"volume": "1",
"year": "2023"
},
{
"DOI": "10.1016/j.bmc.2020.115349",
"doi-asserted-by": "crossref",
"key": "ref_33",
"unstructured": "Samuels, E.R., and Sevrioukova, I.F. (2020). An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4. Bioorganic Med. Chem., 28."
},
{
"DOI": "10.1021/acs.biochem.7b00334",
"article-title": "High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4",
"author": "Sevrioukova",
"doi-asserted-by": "crossref",
"first-page": "3058",
"journal-title": "Biochemistry",
"key": "ref_34",
"volume": "56",
"year": "2017"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "crossref",
"first-page": "455",
"journal-title": "J. Comput. Chem.",
"key": "ref_35",
"volume": "31",
"year": "2010"
}
],
"reference-count": 35,
"references-count": 35,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2813-2998/4/4/50"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "In Silico Evaluation of Structural Consequences in the Human CYP3A4 Caused by Molnupiravir-Induced Mutations During COVID-19 Treatment",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "4"
}