Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)
et al., International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.138, Nov 2020
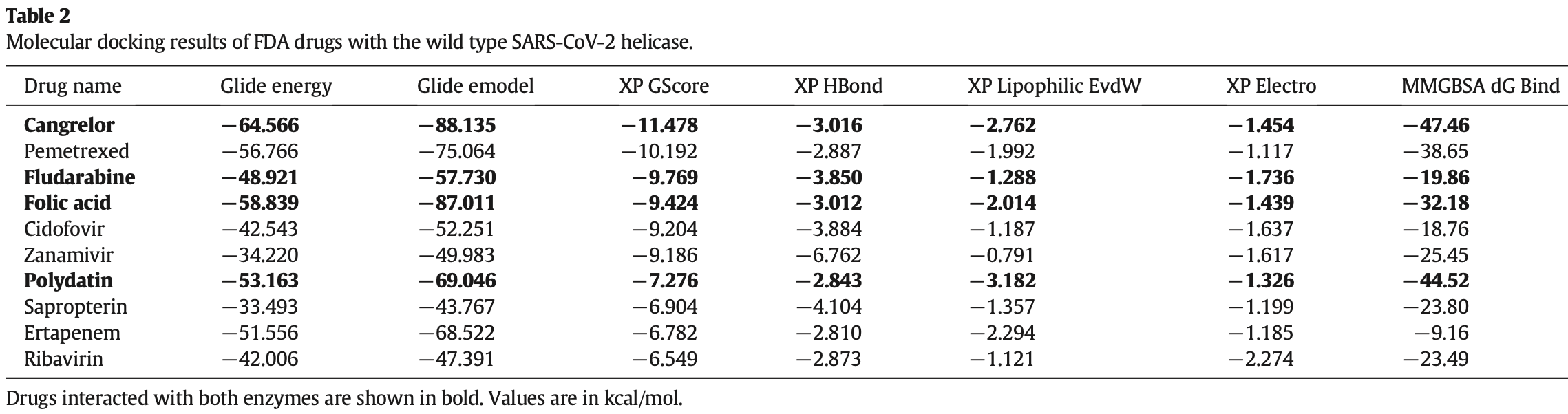
In silico study reporting that folic acid, cangrelor, fludarabine, and polydatin are potential inhibitors of SARS-CoV-2 Nsp13.
14 preclinical studies support the efficacy of vitamin B9 for COVID-19:
Vitamin B9 has been identified by the European Food Safety Authority (EFSA) as having sufficient evidence for a causal relationship between intake and optimal immune system function12-14.
Vitamin B9 inhibits SARS-CoV-2 in silico3-11, reduces spike protein binding ability11, binds with the spike protein receptor binding domain for alpha and omicron variants2, inhibits the SARS-CoV-2 nucleocapsid protein3, inhibits 3CLpro and PLpro in enzymatic assays2, significantly reduces infection for alpha and omicron SARS-CoV-2 pseudoviruses2, and inhibits ACE2 expression and SARS-CoV-2 infection in a mouse model11.
1.
Wu et al., Biomarkers Prediction and Immune Landscape in Covid-19 and “Brain Fog”, Elsevier BV, doi:10.2139/ssrn.4897774.
2.
Pennisi et al., An Integrated In Silico and In Vitro Approach for the Identification of Natural Products Active against SARS-CoV-2, Biomolecules, doi:10.3390/biom14010043.
3.
Chen et al., Folic acid: a potential inhibitor against SARS-CoV-2 nucleocapsid protein, Pharmaceutical Biology, doi:10.1080/13880209.2022.2063341.
4.
Eskandari, V., Repurposing the natural compounds as potential therapeutic agents for COVID-19 based on the molecular docking study of the main protease and the receptor-binding domain of spike protein, Journal of Molecular Modeling, doi:10.1007/s00894-022-05138-3.
5.
Pandya et al., Unravelling Vitamin B12 as a potential inhibitor against SARS-CoV-2: A computational approach, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2022.100951.
6.
Serseg et al., Hispidin and Lepidine E: Two Natural Compounds and Folic Acid as Potential Inhibitors of 2019-novel Coronavirus Main Protease (2019-nCoVMpro), Molecular Docking and SAR Study, Current Computer-Aided Drug Design, doi:10.2174/1573409916666200422075440.
7.
Hosseini et al., Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs, Precision Clinical Medicine, doi:10.1093/pcmedi/pbab001.
8.
Ugurel et al., Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13), International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.138.
9.
Kumar et al., In silico virtual screening-based study of nutraceuticals predicts the therapeutic potentials of folic acid and its derivatives against COVID-19, VirusDisease, doi:10.1007/s13337-020-00643-6.
10.
Moatasim et al., Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E, Microorganisms, doi:10.3390/microorganisms11112777.
11.
Zhang et al., Folic acid restricts SARS-CoV-2 invasion by methylating ACE2, Frontiers in Microbiology, doi:10.3389/fmicb.2022.980903.
12.
Galmés et al., Suboptimal Consumption of Relevant Immune System Micronutrients Is Associated with a Worse Impact of COVID-19 in Spanish Populations, Nutrients, doi:10.3390/nu14112254.
13.
Galmés (B) et al., Current State of Evidence: Influence of Nutritional and Nutrigenetic Factors on Immunity in the COVID-19 Pandemic Framework, Nutrients, doi:10.3390/nu12092738.
14.
EFSA, Scientific Opinion on the substantiation of health claims related to folate and blood formation (ID 79), homocysteine metabolism (ID 80), energy-yielding metabolism (ID 90), function of the immune system (ID 91), function of blood vessels (ID 94, 175, 192), cell division (ID 193), and maternal tissue growth during pregnancy (ID 2882) pursuant to Article 13(1) of Regulation (EC) No 1924/2006, EFSA Journal, doi:10.2903/j.efsa.2009.1213.
Ugurel et al., 30 Nov 2020, peer-reviewed, 8 authors.
Contact: dilekbalik@gmail.com, dbalik@yildiz.edu.tr.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)
International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.138
SARS-CoV-2 has caused COVID-19 outbreak with nearly 2 M infected people and over 100K death worldwide, until middle of April 2020. There is no confirmed drug for the treatment of COVID-19 yet. As the disease spread fast and threaten human life, repositioning of FDA approved drugs may provide fast options for treatment. In this aspect, structure-based drug design could be applied as a powerful approach in distinguishing the viral drug target regions from the host. Evaluation of variations in SARS-CoV-2 genome may ease finding specific drug targets in the viral genome. In this study, 3458 SARS-CoV-2 genome sequences isolated from all around the world were analyzed. Incidence of C17747T and A17858G mutations were observed to be much higher than others and they were on Nsp13, a vital enzyme of SARS-CoV-2. Effect of these mutations was evaluated on protein-drug interactions using in silico methods. The most potent drugs were found to interact with the key and neighbor residues of the active site responsible from ATP hydrolysis. As result, cangrelor, fludarabine, folic acid and polydatin were determined to be the most potent drugs which have potency to inhibit both the wild type and mutant SARS-CoV-2 helicase. Clinical data supporting these findings would be important towards overcoming COVID-19.
References
Ai, Zhang, Zhang, Xu, Zhang, Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned, Emerg. Microbes Infect, doi:10.1080/22221751.2020.1738905
Barnes, Gray, Bioinformatics for Geneticists
Beck, Zhu, Oliveira, Smith, Rich et al., Mechanism of action of methotrexate against zika virus, Viruses, doi:10.3390/v11040338
Benson, Karsch-Mizrachi, Lipman, Ostell, Wheeler, None, Nucleic Acids Res, doi:10.1093/nar/gkg057
Berendsen, Postma, Van Gunsteren, Dinola, Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys, doi:10.1063/1.448118
Borowski, Lang, Niebuhr, Haag, Schmitz et al., Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP), Acta Biochim. Pol, doi:10.18388/abp.2001_3908
Borowski, Mueller, Niebuhr, Kalitzky, Hwang et al., ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy, Acta Biochim. Pol, doi:10.18388/abp.2000_4075
Briguglio, Piras, Corona, Carta, Inhibition of RNA helicases of ssRNA + virus belonging to Flaviviridae, Coronaviridae and Picornaviridae families, Int. J. Med. Chem, doi:10.1155/2011/213135
Cada, Baker, Ingram, None, Hosp. Pharm, doi:10.1310/hpj5010-922
Case, Darden, Cheatham, Simmerling, Wang et al., Amber 12 Reference Manual
Colovos, Yeates, Verification of protein structures: patterns of nonbonded atomic interactions, Protein Sci, doi:10.1002/pro.5560020916
Du, Peng, Zhang, Polydatin: a review of pharmacology and pharmacokinetics, Pharm. Biol, doi:10.3109/13880209.2013.792849
Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res, doi:10.1093/nar/gkh340
Elbe, Buckland-Merrett, Data, disease and diplomacy: GISAID's innovative contribution to global health, Glob. Challenges, doi:10.1002/gch2.1018
Elfiky, Anti-HCV, nucleotide inhibitors, repurposing against COVID-19, Life Sci, doi:10.1016/j.lfs.2020.117477
Eswar, Webb, Sali, Comparative protein structure modeling using MODEL-LER, Curr. Protoc. Bioinforma, doi:10.1002/cpbi.3
Fairman-Williams, Guenther, Jankowsky, SF1 and SF2: family matters, Curr. Opin. Struct. Biol, doi:10.1016/j.sbi.2010.03.011.SF1
Fang, Chen, Tay, Ng, Liu, An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells, Virology, doi:10.1016/j.virol.2006.08.020
Fletcher, Powell, A rapidly convergent descent method for minimization, Comput. J, doi:10.1093/comjnl/6.2.163
Fletcher, Reeves, Function minimization by conjugate gradients, Comput. J, doi:10.1093/comjnl/7.2.149
Fraternale, Casabianca, Tonelli, Vallanti, Chiarantini et al., Inhibition of murine AIDS by alternate administration of azidothymidine and fludarabine monophosphate, JAIDS J. Acquir. Immune Defic. Syndr
Frick, Lam, Understanding helicases as a means of virus control, Curr. Pharm. Des, doi:10.2174/138161206776361147
Gilbert, Knight, Biochemistry and clinical applications of ribavirin, Antimicrob. Agents Chemother, doi:10.1128/aac.30.2.201
Gorbalenya, Koonin, Helicases: amino acid sequence comparisons and structure-function relationships, Curr. Opin. Struct. Biol, doi:10.1016/S0959-440X(05)80116-2
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing, doi:10.1101/2020.03.22.002386
Hanna, Shepherd, Fossella, Pereira, Demarinis et al., Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy, J. Clin. Oncol, doi:10.1200/JCO.2004.08.163
Hitchcock, Jaffe, Martin, Stagg, Cidofovir, a new agent with potent anti-herpesvirus activity, Antivir. Chem. Chemother, doi:10.1177/095632029600700301
Huennekens, Folic acid coenzymes in the biosynthesis of purines and pyrimidines, Vitam. Horm, doi:10.1016/s0083-6729(08)60762-1
Jang, Lee, Yeo, Jeong, Kim, Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase, Biochem. Biophys. Res. Commun, doi:10.1016/j.bbrc.2007.12.020
Jia, Yan, Ren, Wu, Wang et al., Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis, Nucleic Acids Res, doi:10.1093/nar/gkz409
Jorgensen, Chandrasekhar, Madura, Impey, Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys, doi:10.1063/1.445869
Katoh, Rozewicki, Yamada, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization, Brief. Bioinform, doi:10.1093/bib/bbx108
Keating, O'brien, Lerner, Koller, Beran et al., Long-term follow-up of patients with chronic lymphocytic leukemia (CLL) receiving fludarabine regimens as initial therapy, Blood, doi:10.1182/blood.V92.4.1165
Kuraku, Zmasek, Nishimura, Katoh, aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity, Nucleic Acids Res, doi:10.1093/nar/gkt389
Kwong, Rao, Jeang, Viral and cellular RNA helicases as antiviral targets, Nat. Rev. Drug Discov, doi:10.1038/nrd1853
Li, Wang, Xu, Cao, Potential antiviral therapeutics for 2019 novel coronavirus, Zhonghua Jie He He Hu Xi Za Zhi, doi:10.3760/cma.j.issn.1001-0939.2020.0002
Lin, Ho, Chuo, Li, Wang et al., Effective inhibition of MERS-CoV infection by resveratrol, BMC Infect. Dis, doi:10.1186/s12879-017-2253-8
Lovell, Davis, Adrendall, De Bakker, Word et al., Structure validation by C alpha geomF, Proteins-Structure Funct. Genet, doi:10.1002/prot.10286
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736(20)30251-8
Luthy, Bowei, Einsenberg, Verify3D: assessment of protein models with three-dimensional profiles, Methods Enzymol
Maier, Martinez, Kasavajhala, Wickstrom, Hauser et al., ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB, J. Chem. Theory Comput, doi:10.1021/acs.jctc.5b00255
Martin, Jensen, Ribavirin in the treatment of chronic hepatitis C, J. Gastroenterol. Hepatol, doi:10.1111/j.1440-1746.2008.05398.x
Mirza, Froeyen, Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 RNA-dependent RNA polymerase and Nsp13 helicase, doi:10.20944/preprints202003.0085.v1
Moinpour, Barker, Guzman, Jewett, Langlais et al., Determining Protein Structure by Tyrosine Bioconjugation
Morgan, Rubenstein, Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome, PLoS One, doi:10.1371/journal.pone.0053785
O'leary, Wright, Brister, Ciufo, Haddad et al., Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation, Nucleic Acids Res, doi:10.1093/nar/gkv1189
Papich, Mark, Dmv, Saunders Handbook of Veterinary Drugs-e-book: Small and Large Animal
Parvez, Subbarao, Molecular analysis and modeling of hepatitis e virus helicase and identification of novel inhibitors by virtual screening, Biomed. Res. Int, doi:10.1155/2018/5753804
Pastor, Brooks, Szabo, An analysis of the accuracy of Langevin and molecular dynamics algorithms, Mol. Phys, doi:10.1080/00268978800101881
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera -a visualization system for exploratory research and analysis, J. Comput. Chem, doi:10.1002/jcc.20084
Rai, Peterson, Appelbaum, Kolitz, Elias et al., Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia, N. Engl. J. Med, doi:10.1056/NEJM200012143432402
Roe, Cheatham, PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data, J. Chem. Theory Comput, doi:10.1021/ct400341p
Rusthoven, Eisenhauer, Butts, Gregg, Dancey et al., Multitargeted antifolate LY231514 as first-line chemotherapy for patients with advanced non-small-cell lung cancer: a phase II study, J. Clin. Oncol
Ryckaert, Ciccotti, Berendsen, Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of nalkanes, J. Comput. Phys, doi:10.1016/0021-9991(77)90098-5
Sanford, Keating, None, Sapropterin, doi:10.2165/00003495-200969040-00006
Saw, Pan, Subramanian Manimekalai, Grüber, Structural features of Zika virus non-structural proteins 3 and -5 and its individual domains in solution as well as insights into NS3 inhibition, Antivir. Res, doi:10.1016/j.antiviral.2017.02.005
Sevier, Kaiser, Formation and transfer of disulphide bonds in living cells, Nat. Rev. Mol. Cell Biol
Shum, Tanner, Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase, Chembiochem, doi:10.1002/cbic.200800491
Snijder, Decroly, Ziebuhr, The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing, doi:10.1016/bs.aivir.2016.08.008
Srinivasan, Cui, Gao, Liu, Lu et al., Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins, Viruses
Stadler, Demmel, Ollivier, Seydel, Picosecond to nanosecond dynamics provide a source of conformational entropy for protein folding, Phys. Chem. Chem. Phys, doi:10.1039/c6cp04146a
Sterling, Irwin, ZINC 15 -ligand discovery for everyone, J. Chem. Inf. Model, doi:10.1021/acs.jcim.5b00559
Subissi, Imbert, Ferron, Collet, Coutard et al., SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets, Antivir. Res, doi:10.1016/j.antiviral.2013.11.006
Vyas, Ukawala, Ghate, Chintha, Homology modeling a fast tool for drug discovery: current perspectives, Indian J. Pharm. Sci, doi:10.4103/0250-474X.102537
Wallner, Elofsson, Can correct protein models be identified?, Protein Sci, doi:10.1110/ps.0236803
Wang, Huang, Gao, Chen, Xinxie et al., Screening anti-TMV agents targeting tobacco mosaic virus helicase protein, Pestic. Biochem. Physiol, doi:10.1016/j.pestbp.2019.07.017
Wang, Huang, Wong, Wang, Zhang et al., On the mechanisms of bananin activity against severe acute respiratory syndrome coronavirus, FEBS J, doi:10.1111/j.1742-4658.2010.07961.x
Wang, Liu, Enterovirus 71: epidemiology, pathogenesis and management, Expert Rev. Anti-Infect. Ther, doi:10.1586/eri.09.45
Wang, Wang, Zhou, Deng, Li et al., Leucovorin enhances the anti-cancer effect of bortezomib in colorectal cancer cells, Sci. Rep, doi:10.1038/s41598-017-00839-9
Waterhouse, Bertoni, Bienert, Studer, Tauriello et al., SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res, doi:10.1093/nar/gky427
Waterhouse, Procter, Martin, Clamp, Barton, Jalview Version 2 -a multiple sequence alignment editor and analysis workbench, Bioinformatics, doi:10.1093/bioinformatics/btp033
Waye, Sing, Anti-viral drugs for human adenoviruses, Pharmaceuticals, doi:10.3390/ph3103343
Wiederstein, Sippl, ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins, Nucleic Acids Res, doi:10.1093/nar/gkm290
Williams, Kelley, Gnuplot 4.5: An Interactive Plotting Program
Wu, Liu, Yang, Zhang, Zhong et al., Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharm. Sin. B, doi:10.1016/j.apsb.2020.02.008
Wu, Peng, Huang, Ding, Wang et al., Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China, Cell Host Microbe, doi:10.1016/j.chom.2020.02.001
Wu, Zhao, Yu, Chen, Wang et al., A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Yano, Kohno, Ohkusa, Mochizuki, Yamada et al., Effect of milrinone on left ventricular relaxation and Ca2+ uptake function of cardiac sarcoplasmic reticulum, Am. J. Physiol. Circ. Physiol, doi:10.1152/ajpheart.2000.279.4.H1898
Yin, Wunderink, MERS, SARS and other coronaviruses as causes of pneumonia, Respirology, doi:10.1111/resp.13196
Yu, Lee, Lee, Kim, Chin et al., Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13, Bioorganic Med. Chem. Lett, doi:10.1016/j.bmcl.2012.04.081
Zhang, Li, Gu, Wang, Shi et al., Resveratrol inhibits enterovirus 71 replication and pro-inflammatory cytokine secretion in rhabdosarcoma cells through blocking IKKs/NF-κB signaling pathway, PLoS One, doi:10.1371/journal.pone.0116879
Zhang, Liu, Potential interventions for novel coronavirus in China: a systematic review, J. Med. Virol, doi:10.1002/jmv.25707
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zhu, Zhang, Wang, Li, Yang et al., A novel coronavirus from patients with pneumonia in China, 2019, N. Engl. J. Med, doi:10.1056/NEJMoa2001017
DOI record:
{
"DOI": "10.1016/j.ijbiomac.2020.09.138",
"ISSN": [
"0141-8130"
],
"URL": "http://dx.doi.org/10.1016/j.ijbiomac.2020.09.138",
"alternative-id": [
"S0141813020344974"
],
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "International Journal of Biological Macromolecules"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.ijbiomac.2020.09.138"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2020 Elsevier B.V. All rights reserved."
}
],
"author": [
{
"affiliation": [],
"family": "Ugurel",
"given": "Osman Mutluhan",
"sequence": "first"
},
{
"affiliation": [],
"family": "Mutlu",
"given": "Ozal",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sariyer",
"given": "Emrah",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kocer",
"given": "Sinem",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ugurel",
"given": "Erennur",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Inci",
"given": "Tugba Gul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ata",
"given": "Oguz",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Turgut-Balik",
"given": "Dilek",
"sequence": "additional"
}
],
"container-title": "International Journal of Biological Macromolecules",
"container-title-short": "International Journal of Biological Macromolecules",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2020,
9,
24
]
],
"date-time": "2020-09-24T16:31:43Z",
"timestamp": 1600965103000
},
"deposited": {
"date-parts": [
[
2020,
12,
18
]
],
"date-time": "2020-12-18T23:50:33Z",
"timestamp": 1608335433000
},
"indexed": {
"date-parts": [
[
2023,
3,
27
]
],
"date-time": "2023-03-27T23:28:39Z",
"timestamp": 1679959719479
},
"is-referenced-by-count": 24,
"issued": {
"date-parts": [
[
2020,
11
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
11,
1
]
],
"date-time": "2020-11-01T00:00:00Z",
"timestamp": 1604188800000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0141813020344974?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0141813020344974?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "1687-1696",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2020,
11
]
]
},
"published-print": {
"date-parts": [
[
2020,
11
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1016/j.chom.2020.02.001",
"article-title": "Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "325",
"journal-title": "Cell Host Microbe",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0005",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A pneumonia outbreak associated with a new coronavirus of probable bat origin",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nature",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0010",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2001017",
"article-title": "A novel coronavirus from patients with pneumonia in China, 2019",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "727",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0020",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1080/22221751.2020.1738905",
"article-title": "Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned",
"author": "Ai",
"doi-asserted-by": "crossref",
"first-page": "597",
"journal-title": "Emerg. Microbes Infect.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0030",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"article-title": "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "565",
"journal-title": "Lancet",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0035",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"article-title": "A new coronavirus associated with human respiratory disease in China",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "265",
"journal-title": "Nature",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0040",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2007.12.020",
"article-title": "Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase",
"author": "Jang",
"doi-asserted-by": "crossref",
"first-page": "738",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0045",
"volume": "366",
"year": "2008"
},
{
"article-title": "Inhibition of RNA helicases of ssRNA + virus belonging to Flaviviridae, Coronaviridae and Picornaviridae families",
"author": "Briguglio",
"first-page": "1",
"journal-title": "Int. J. Med. Chem.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0050",
"volume": "2011",
"year": "2011"
},
{
"DOI": "10.2174/138161206776361147",
"article-title": "Understanding helicases as a means of virus control",
"author": "Frick",
"doi-asserted-by": "crossref",
"first-page": "1315",
"journal-title": "Curr. Pharm. Des.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0055",
"volume": "12",
"year": "2006"
},
{
"author": "Snijder",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0060",
"series-title": "The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing",
"year": "2016"
},
{
"DOI": "10.1038/nrd1853",
"article-title": "Viral and cellular RNA helicases as antiviral targets",
"author": "Kwong",
"doi-asserted-by": "crossref",
"first-page": "845",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0065",
"volume": "4",
"year": "2005"
},
{
"DOI": "10.1002/cbic.200800491",
"article-title": "Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase",
"author": "Shum",
"doi-asserted-by": "crossref",
"first-page": "3037",
"journal-title": "Chembiochem",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0070",
"volume": "9",
"year": "2008"
},
{
"DOI": "10.1016/S0959-440X(05)80116-2",
"article-title": "Helicases: amino acid sequence comparisons and structure-function relationships",
"author": "Gorbalenya",
"doi-asserted-by": "crossref",
"first-page": "419",
"journal-title": "Curr. Opin. Struct. Biol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0075",
"volume": "3",
"year": "1993"
},
{
"DOI": "10.1016/j.sbi.2010.03.011",
"article-title": "SF1 and SF2: family matters",
"author": "Fairman-Williams",
"doi-asserted-by": "crossref",
"first-page": "313",
"journal-title": "Curr. Opin. Struct. Biol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0080",
"volume": "20",
"year": "2010"
},
{
"DOI": "10.1016/j.antiviral.2013.11.006",
"article-title": "SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets",
"author": "Subissi",
"doi-asserted-by": "crossref",
"first-page": "122",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0085",
"volume": "101",
"year": "2014"
},
{
"DOI": "10.1093/nar/gkz409",
"article-title": "Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis",
"author": "Jia",
"doi-asserted-by": "crossref",
"first-page": "6538",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0090",
"volume": "47",
"year": "2019"
},
{
"author": "Mirza",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0095",
"series-title": "Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 RNA-dependent RNA polymerase and Nsp13 helicase",
"year": "2020"
},
{
"DOI": "10.1016/j.bmcl.2012.04.081",
"article-title": "Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13",
"author": "Yu",
"doi-asserted-by": "crossref",
"first-page": "4049",
"journal-title": "Bioorganic Med. Chem. Lett.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0100",
"volume": "22",
"year": "2012"
},
{
"DOI": "10.1016/j.virol.2006.08.020",
"article-title": "An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells",
"author": "Fang",
"doi-asserted-by": "crossref",
"first-page": "136",
"journal-title": "Virology",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0105",
"volume": "358",
"year": "2007"
},
{
"DOI": "10.1111/j.1742-4658.2010.07961.x",
"article-title": "On the mechanisms of bananin activity against severe acute respiratory syndrome coronavirus",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "383",
"journal-title": "FEBS J.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0110",
"volume": "278",
"year": "2011"
},
{
"DOI": "10.1093/nar/gkv1189",
"article-title": "Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation",
"author": "O’Leary",
"doi-asserted-by": "crossref",
"first-page": "D733",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0115",
"volume": "44",
"year": "2015"
},
{
"DOI": "10.1002/gch2.1018",
"article-title": "Data, disease and diplomacy: GISAID’s innovative contribution to global health",
"author": "Elbe",
"doi-asserted-by": "crossref",
"first-page": "33",
"journal-title": "Glob. Challenges.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0125",
"volume": "1",
"year": "2017"
},
{
"DOI": "10.1093/nar/gkg057",
"article-title": "GenBank",
"author": "Benson",
"doi-asserted-by": "crossref",
"first-page": "23",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0130",
"volume": "31",
"year": "2003"
},
{
"DOI": "10.1093/nar/gkt389",
"article-title": "aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity",
"author": "Kuraku",
"doi-asserted-by": "crossref",
"first-page": "W22",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0135",
"volume": "41",
"year": "2013"
},
{
"DOI": "10.1093/bib/bbx108",
"article-title": "MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization",
"author": "Katoh",
"doi-asserted-by": "crossref",
"first-page": "1160",
"journal-title": "Brief. Bioinform.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0140",
"volume": "20",
"year": "2017"
},
{
"DOI": "10.1093/bioinformatics/btp033",
"article-title": "Jalview Version 2—a multiple sequence alignment editor and analysis workbench",
"author": "Waterhouse",
"doi-asserted-by": "crossref",
"first-page": "1189",
"journal-title": "Bioinformatics",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0145",
"volume": "25",
"year": "2009"
},
{
"DOI": "10.1093/nar/gkh340",
"article-title": "MUSCLE: multiple sequence alignment with high accuracy and high throughput",
"author": "Edgar",
"doi-asserted-by": "crossref",
"first-page": "1792",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0150",
"volume": "32",
"year": "2004"
},
{
"article-title": "Comparative protein structure modeling using MODELLER",
"author": "Eswar",
"first-page": "1",
"issue": "6",
"journal-title": "Curr. Protoc. Bioinforma.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0160",
"volume": "5",
"year": "2006"
},
{
"DOI": "10.1093/nar/gky427",
"article-title": "SWISS-MODEL: homology modelling of protein structures and complexes",
"author": "Waterhouse",
"doi-asserted-by": "crossref",
"first-page": "W296",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0165",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.1093/nar/gkm290",
"article-title": "ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins",
"author": "Wiederstein",
"doi-asserted-by": "crossref",
"first-page": "407",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0170",
"volume": "35",
"year": "2007"
},
{
"DOI": "10.1002/pro.5560020916",
"article-title": "Verification of protein structures: patterns of nonbonded atomic interactions",
"author": "Colovos",
"doi-asserted-by": "crossref",
"first-page": "1511",
"journal-title": "Protein Sci.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0175",
"volume": "2",
"year": "1993"
},
{
"DOI": "10.1016/S0076-6879(97)77022-8",
"article-title": "Verify3D: assessment of protein models with three-dimensional profiles",
"author": "Luthy",
"doi-asserted-by": "crossref",
"first-page": "396",
"journal-title": "Methods Enzymol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0180",
"volume": "277",
"year": "1997"
},
{
"article-title": "Basic local alignment search tool",
"author": "Lovell",
"first-page": "437",
"issue": "2003",
"journal-title": "Journal of Molecular Biology.etry: phi,psi and C beta deviation, Proteins-Structure Funct. Genet",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0185",
"volume": "50",
"year": "1990"
},
{
"DOI": "10.1110/ps.0236803",
"article-title": "Can correct protein models be identified?",
"author": "Wallner",
"doi-asserted-by": "crossref",
"first-page": "1073",
"journal-title": "Protein Sci.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0190",
"volume": "12",
"year": "2003"
},
{
"DOI": "10.1002/jcc.20084",
"article-title": "UCSF Chimera - a visualization system for exploratory research and analysis",
"author": "Pettersen",
"doi-asserted-by": "crossref",
"first-page": "1605",
"journal-title": "J. Comput. Chem.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0195",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1039/C6CP04146A",
"article-title": "Picosecond to nanosecond dynamics provide a source of conformational entropy for protein folding",
"author": "Stadler",
"doi-asserted-by": "crossref",
"first-page": "21527",
"journal-title": "Phys. Chem. Chem. Phys.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0200",
"volume": "18",
"year": "2016"
},
{
"DOI": "10.4103/0250-474X.102537",
"article-title": "Homology modeling a fast tool for drug discovery: current perspectives",
"author": "Vyas",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Indian J. Pharm. Sci.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0205",
"volume": "74",
"year": "2012"
},
{
"author": "Case",
"first-page": "350",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0210",
"series-title": "Amber 12 Reference Manual",
"year": "2012"
},
{
"DOI": "10.1021/acs.jctc.5b00255",
"article-title": "ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB",
"author": "Maier",
"doi-asserted-by": "crossref",
"first-page": "3696",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0215",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1063/1.445869",
"article-title": "Comparison of simple potential functions for simulating liquid water",
"author": "Jorgensen",
"doi-asserted-by": "crossref",
"first-page": "926",
"journal-title": "J. Chem. Phys.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0220",
"volume": "79",
"year": "1983"
},
{
"DOI": "10.1093/comjnl/6.2.163",
"article-title": "A rapidly convergent descent method for minimization",
"author": "Fletcher",
"doi-asserted-by": "crossref",
"first-page": "163",
"journal-title": "Comput. J.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0225",
"volume": "6",
"year": "1963"
},
{
"DOI": "10.1093/comjnl/7.2.149",
"article-title": "Function minimization by conjugate gradients",
"author": "Fletcher",
"doi-asserted-by": "crossref",
"first-page": "149",
"journal-title": "Comput. J.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0230",
"volume": "7",
"year": "1964"
},
{
"DOI": "10.1080/00268978800101881",
"article-title": "An analysis of the accuracy of Langevin and molecular dynamics algorithms",
"author": "Pastor",
"doi-asserted-by": "crossref",
"first-page": "1409",
"journal-title": "Mol. Phys.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0235",
"volume": "65",
"year": "1988"
},
{
"DOI": "10.1063/1.448118",
"article-title": "Molecular dynamics with coupling to an external bath",
"author": "Berendsen",
"doi-asserted-by": "crossref",
"first-page": "3684",
"journal-title": "J. Chem. Phys.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0240",
"volume": "81",
"year": "1984"
},
{
"DOI": "10.1016/0021-9991(77)90098-5",
"article-title": "Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes",
"author": "Ryckaert",
"doi-asserted-by": "crossref",
"first-page": "327",
"journal-title": "J. Comput. Phys.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0245",
"volume": "23",
"year": "1977"
},
{
"DOI": "10.1021/ct400341p",
"article-title": "PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data",
"author": "Roe",
"doi-asserted-by": "crossref",
"first-page": "3084",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0250",
"volume": "9",
"year": "2013"
},
{
"key": "10.1016/j.ijbiomac.2020.09.138_bb0255",
"unstructured": "T. Williams and Kelley, C., Gnuplot 4.5: An Interactive Plotting Program, (n.d.)."
},
{
"DOI": "10.1021/acs.jcim.5b00559",
"article-title": "ZINC 15 – ligand discovery for everyone",
"author": "Sterling",
"doi-asserted-by": "crossref",
"first-page": "2324",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0260",
"volume": "55",
"year": "2015"
},
{
"DOI": "10.1016/j.apsb.2020.02.008",
"article-title": "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods",
"author": "Wu",
"doi-asserted-by": "crossref",
"journal-title": "Acta Pharm. Sin. B",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0265",
"year": "2020"
},
{
"DOI": "10.3390/v12040360",
"article-title": "Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins",
"author": "Srinivasan",
"doi-asserted-by": "crossref",
"first-page": "360",
"journal-title": "Viruses",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0270",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1371/journal.pone.0053785",
"article-title": "Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome",
"author": "Morgan",
"doi-asserted-by": "crossref",
"first-page": "e53785",
"issue": "1",
"journal-title": "PLoS One",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0275",
"volume": "8",
"year": "2013"
},
{
"author": "Barnes",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0280",
"series-title": "Bioinformatics for Geneticists",
"year": "2003"
},
{
"author": "Moinpour",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0285",
"series-title": "Determining Protein Structure by Tyrosine Bioconjugation, BioRxiv",
"year": "2020"
},
{
"DOI": "10.1038/nrm954",
"article-title": "Formation and transfer of disulphide bonds in living cells",
"author": "Sevier",
"doi-asserted-by": "crossref",
"first-page": "836",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0290",
"volume": "3",
"year": "2002"
},
{
"DOI": "10.1016/S0083-6729(08)60762-1",
"article-title": "Folic acid coenzymes in the biosynthesis of purines and pyrimidines",
"author": "Huennekens",
"doi-asserted-by": "crossref",
"first-page": "375",
"journal-title": "Vitam. Horm.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0295",
"volume": "26",
"year": "1968"
},
{
"DOI": "10.1310/hpj5010-922",
"article-title": "Cangrelor",
"author": "Cada",
"doi-asserted-by": "crossref",
"first-page": "922",
"journal-title": "Hosp. Pharm.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0300",
"volume": "50",
"year": "2015"
},
{
"DOI": "10.1056/NEJM200012143432402",
"article-title": "Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia",
"author": "Rai",
"doi-asserted-by": "crossref",
"first-page": "1750",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0305",
"volume": "343",
"year": "2000"
},
{
"DOI": "10.1182/blood.V92.4.1165",
"article-title": "Long-term follow-up of patients with chronic lymphocytic leukemia (CLL) receiving fludarabine regimens as initial therapy",
"author": "Keating",
"doi-asserted-by": "crossref",
"first-page": "1165",
"journal-title": "Blood.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0310",
"volume": "92",
"year": "1998"
},
{
"DOI": "10.1097/00042560-200003010-00001",
"article-title": "Inhibition of murine AIDS by alternate administration of azidothymidine and fludarabine monophosphate",
"author": "Fraternale",
"doi-asserted-by": "crossref",
"first-page": "209",
"journal-title": "JAIDS J. Acquir. Immune Defic. Syndr.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0315",
"volume": "23",
"year": "2000"
},
{
"DOI": "10.3109/13880209.2013.792849",
"article-title": "Polydatin: a review of pharmacology and pharmacokinetics",
"author": "Du",
"doi-asserted-by": "crossref",
"first-page": "1347",
"journal-title": "Pharm. Biol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0320",
"volume": "51",
"year": "2013"
},
{
"article-title": "Resveratrol inhibits enterovirus 71 replication and pro-inflammatory cytokine secretion in rhabdosarcoma cells through blocking IKKs/NF-κB signaling pathway",
"author": "Zhang",
"first-page": "1",
"journal-title": "PLoS One",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0325",
"volume": "10",
"year": "2015"
},
{
"DOI": "10.1186/s12879-017-2253-8",
"article-title": "Effective inhibition of MERS-CoV infection by resveratrol",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "BMC Infect. Dis.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0330",
"volume": "17",
"year": "2017"
},
{
"DOI": "10.1016/j.antiviral.2017.02.005",
"article-title": "Structural features of Zika virus non-structural proteins 3 and -5 and its individual domains in solution as well as insights into NS3 inhibition",
"author": "Saw",
"doi-asserted-by": "crossref",
"first-page": "73",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0335",
"volume": "141",
"year": "2017"
},
{
"DOI": "10.3390/ph3103343",
"article-title": "Anti-viral drugs for human adenoviruses",
"author": "Waye",
"doi-asserted-by": "crossref",
"first-page": "3343",
"journal-title": "Pharmaceuticals",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0340",
"volume": "3",
"year": "2010"
},
{
"DOI": "10.1200/JCO.1999.17.4.1194",
"article-title": "Multitargeted antifolate LY231514 as first-line chemotherapy for patients with advanced non–small-cell lung cancer: a phase II study",
"author": "Rusthoven",
"doi-asserted-by": "crossref",
"first-page": "1194",
"journal-title": "J. Clin. Oncol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0345",
"volume": "17",
"year": "1999"
},
{
"DOI": "10.1200/JCO.2004.08.163",
"article-title": "Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy",
"author": "Hanna",
"doi-asserted-by": "crossref",
"first-page": "1589",
"journal-title": "J. Clin. Oncol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0350",
"volume": "22",
"year": "2004"
},
{
"DOI": "10.1177/095632029600700301",
"article-title": "Cidofovir, a new agent with potent anti-herpesvirus activity",
"author": "Hitchcock",
"doi-asserted-by": "crossref",
"first-page": "115",
"journal-title": "Antivir. Chem. Chemother.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0355",
"volume": "7",
"year": "1996"
},
{
"author": "Papich",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0365",
"series-title": "Saunders Handbook of Veterinary Drugs-e-book: Small and Large Animal",
"year": "2015"
},
{
"DOI": "10.2165/00003495-200969040-00006",
"article-title": "Sapropterin",
"author": "Sanford",
"doi-asserted-by": "crossref",
"first-page": "461",
"journal-title": "Drugs",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0370",
"volume": "69",
"year": "2009"
},
{
"DOI": "10.1128/AAC.30.2.201",
"article-title": "Biochemistry and clinical applications of ribavirin",
"author": "Gilbert",
"doi-asserted-by": "crossref",
"first-page": "201",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0375",
"volume": "30",
"year": "1986"
},
{
"DOI": "10.1111/j.1440-1746.2008.05398.x",
"article-title": "Ribavirin in the treatment of chronic hepatitis C",
"author": "Martin",
"doi-asserted-by": "crossref",
"first-page": "844",
"journal-title": "J. Gastroenterol. Hepatol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0380",
"volume": "23",
"year": "2008"
},
{
"DOI": "10.1111/resp.13196",
"article-title": "MERS, SARS and other coronaviruses as causes of pneumonia",
"author": "Yin",
"doi-asserted-by": "crossref",
"first-page": "130",
"journal-title": "Respirology",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0385",
"volume": "23",
"year": "2018"
},
{
"DOI": "10.1002/jmv.25707",
"article-title": "Potential interventions for novel coronavirus in China: a systematic review",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "479",
"journal-title": "J. Med. Virol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0390",
"year": "2020"
},
{
"author": "Gordon",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0395",
"series-title": "A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing, BioRxiv",
"year": "2020"
},
{
"DOI": "10.1016/j.lfs.2020.117477",
"article-title": "Anti-HCV, nucleotide inhibitors, repurposing against COVID-19",
"author": "Elfiky",
"doi-asserted-by": "crossref",
"journal-title": "Life Sci.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0400",
"volume": "248",
"year": "2020"
},
{
"DOI": "10.18388/abp.2000_4075",
"article-title": "ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy",
"author": "Borowski",
"doi-asserted-by": "crossref",
"first-page": "173",
"journal-title": "Acta Biochim. Pol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0405",
"volume": "47",
"year": "2000"
},
{
"DOI": "10.18388/abp.2001_3908",
"article-title": "Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP)",
"author": "Borowski",
"doi-asserted-by": "crossref",
"first-page": "739",
"journal-title": "Acta Biochim. Pol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0410",
"volume": "48",
"year": "2001"
},
{
"article-title": "Screening anti-TMV agents targeting tobacco mosaic virus helicase protein",
"author": "Wang",
"journal-title": "Pestic. Biochem. Physiol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0415",
"year": "2019"
},
{
"DOI": "10.1155/2018/5753804",
"article-title": "Molecular analysis and modeling of hepatitis e virus helicase and identification of novel inhibitors by virtual screening",
"author": "Parvez",
"doi-asserted-by": "crossref",
"journal-title": "Biomed. Res. Int.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0420",
"volume": "2018",
"year": "2018"
},
{
"article-title": "Potential antiviral therapeutics for 2019 novel coronavirus",
"author": "Li",
"journal-title": "Zhonghua Jie He He Hu Xi Za Zhi.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0425",
"volume": "43",
"year": "2020"
},
{
"DOI": "10.3390/v11040338",
"article-title": "Mechanism of action of methotrexate against zika virus",
"author": "Beck",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Viruses.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0430",
"volume": "11",
"year": "2019"
},
{
"article-title": "Leucovorin enhances the anti-cancer effect of bortezomib in colorectal cancer cells",
"author": "Wang",
"first-page": "1",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0435",
"volume": "7",
"year": "2017"
},
{
"DOI": "10.1152/ajpheart.2000.279.4.H1898",
"article-title": "Effect of milrinone on left ventricular relaxation and Ca2+ uptake function of cardiac sarcoplasmic reticulum",
"author": "Yano",
"doi-asserted-by": "crossref",
"first-page": "H1898",
"journal-title": "Am. J. Physiol. Circ. Physiol.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0440",
"volume": "279",
"year": "2000"
},
{
"DOI": "10.1586/eri.09.45",
"article-title": "Enterovirus 71: epidemiology, pathogenesis and management",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "735",
"journal-title": "Expert Rev. Anti-Infect. Ther.",
"key": "10.1016/j.ijbiomac.2020.09.138_bb0445",
"volume": "7",
"year": "2009"
}
],
"reference-count": 84,
"references-count": 84,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0141813020344974"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Molecular Biology",
"General Medicine",
"Biochemistry",
"Structural Biology"
],
"subtitle": [],
"title": "Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1016/elsevier_cm_policy",
"volume": "163"
}