Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs
et al., Precision Clinical Medicine, doi:10.1093/pcmedi/pbab001, Jan 2021
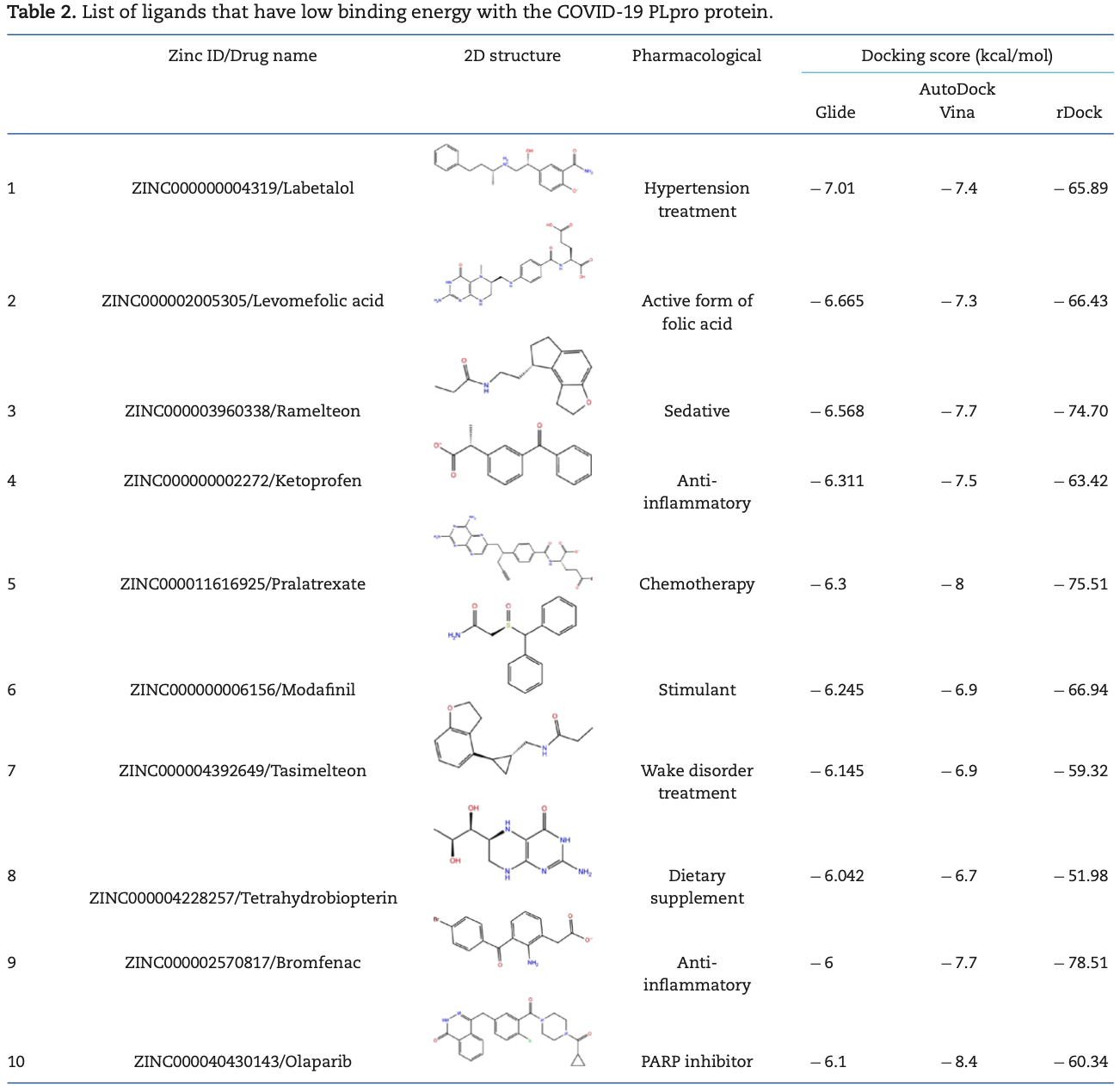
In silico study identifying novel ligands as potential inhibitors for SARS-CoV-2, including antiemetics rolapitant and ondansetron for Mpro; labetalol and levomefolic acid for PLpro; and leucal and antifungal natamycin for RdRp.
14 preclinical studies support the efficacy of vitamin B9 for COVID-19:
Vitamin B9 has been identified by the European Food Safety Authority (EFSA) as having sufficient evidence for a causal relationship between intake and optimal immune system function12-14.
Vitamin B9 inhibits SARS-CoV-2 in silico3-11, reduces spike protein binding ability11, binds with the spike protein receptor binding domain for alpha and omicron variants2, inhibits the SARS-CoV-2 nucleocapsid protein3, inhibits 3CLpro and PLpro in enzymatic assays2, significantly reduces infection for alpha and omicron SARS-CoV-2 pseudoviruses2, and inhibits ACE2 expression and SARS-CoV-2 infection in a mouse model11.
1.
Wu et al., Biomarkers Prediction and Immune Landscape in Covid-19 and “Brain Fog”, Elsevier BV, doi:10.2139/ssrn.4897774.
2.
Pennisi et al., An Integrated In Silico and In Vitro Approach for the Identification of Natural Products Active against SARS-CoV-2, Biomolecules, doi:10.3390/biom14010043.
3.
Chen et al., Folic acid: a potential inhibitor against SARS-CoV-2 nucleocapsid protein, Pharmaceutical Biology, doi:10.1080/13880209.2022.2063341.
4.
Eskandari, V., Repurposing the natural compounds as potential therapeutic agents for COVID-19 based on the molecular docking study of the main protease and the receptor-binding domain of spike protein, Journal of Molecular Modeling, doi:10.1007/s00894-022-05138-3.
5.
Pandya et al., Unravelling Vitamin B12 as a potential inhibitor against SARS-CoV-2: A computational approach, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2022.100951.
6.
Serseg et al., Hispidin and Lepidine E: Two Natural Compounds and Folic Acid as Potential Inhibitors of 2019-novel Coronavirus Main Protease (2019-nCoVMpro), Molecular Docking and SAR Study, Current Computer-Aided Drug Design, doi:10.2174/1573409916666200422075440.
7.
Hosseini et al., Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs, Precision Clinical Medicine, doi:10.1093/pcmedi/pbab001.
8.
Ugurel et al., Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13), International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.138.
9.
Kumar et al., In silico virtual screening-based study of nutraceuticals predicts the therapeutic potentials of folic acid and its derivatives against COVID-19, VirusDisease, doi:10.1007/s13337-020-00643-6.
10.
Moatasim et al., Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E, Microorganisms, doi:10.3390/microorganisms11112777.
11.
Zhang et al., Folic acid restricts SARS-CoV-2 invasion by methylating ACE2, Frontiers in Microbiology, doi:10.3389/fmicb.2022.980903.
12.
Galmés et al., Suboptimal Consumption of Relevant Immune System Micronutrients Is Associated with a Worse Impact of COVID-19 in Spanish Populations, Nutrients, doi:10.3390/nu14112254.
13.
Galmés (B) et al., Current State of Evidence: Influence of Nutritional and Nutrigenetic Factors on Immunity in the COVID-19 Pandemic Framework, Nutrients, doi:10.3390/nu12092738.
14.
EFSA, Scientific Opinion on the substantiation of health claims related to folate and blood formation (ID 79), homocysteine metabolism (ID 80), energy-yielding metabolism (ID 90), function of the immune system (ID 91), function of blood vessels (ID 94, 175, 192), cell division (ID 193), and maternal tissue growth during pregnancy (ID 2882) pursuant to Article 13(1) of Regulation (EC) No 1924/2006, EFSA Journal, doi:10.2903/j.efsa.2009.1213.
Hosseini et al., 18 Jan 2021, peer-reviewed, 4 authors.
Contact: chwang@llu.edu, oxwang@gmail.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs
Precision Clinical Medicine, doi:10.1093/pcmedi/pbab001
The pandemic of novel coronavirus disease 2019 (COVID-19) has rampaged the world, with more than 58.4 million confirmed cases and over 1.38 million deaths across the world by 23 November 2020. There is an urgent need to identify effective drugs and vaccines to fight against the virus. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the family of coronaviruses consisting of four structural and 16 non-structural proteins (NSP). Three non-structural proteins, main protease (Mpro), papain-like protease (PLpro), and RNAdependent RNA polymerase (RdRp), are believed to have a crucial role in replication of the virus. We applied computational ligand-receptor binding modeling and performed comprehensive virtual screening on FDA-approved drugs against these three SARS-CoV-2 proteins using AutoDock Vina, Glide, and rDock. Our computational studies identified six novel ligands as potential inhibitors against SARS-CoV-2, including antiemetics rolapitant and ondansetron for Mpro; labetalol and levomefolic acid for PLpro; and leucal and antifungal natamycin for RdRp. Molecular dynamics simulation confirmed the stability of the ligand-protein complexes. The results of our analysis with some other suggested drugs indicated that chloroquine and hydroxychloroquine had high binding energy (low inhibitory effect) with all three proteins-Mpro, PLpro, and RdRp. In summary, our computational molecular docking approach and virtual screening identified some promising candidate SARS-CoV-2 inhibitors that may be considered for further clinical studies.
Supplementary data Supplementary data are available at PCMEDI online.
Conflict of interest statement All authors declared no conflict of interests. In addition, as an Editorial Board Member of Precision Clinical Medicine, the corresponding author Charles Wang was blinded from reviewing and making decision on this manuscript.
References
Alamri, Altharawi, Alabbas, Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics, Arabian J Chem, doi:10.1016/j.arabjc.2020.08.004
Arul, Kumar, Jeyakanthan, Searching for targetspecific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach, Sci Rep, doi:10.1038/s41598-020-75762-7
Arun, Sharanya, Abhithaj, Drug repurposing to identify therapeutics against COVID 19 with SARS-Cov-2 spike glycoprotein and main protease as targets: an in silico study, ChemRxiv, doi:10.26434/chemrxiv.12090408.v1
Baby, Maity, Mehta, Targeting SARS-CoV-2 RNA-dependent RNA polymerase: An in silico drug repurposing for COVID-19, F1000Research, doi:10.12688/f1000research.26359.1
Baden, Rubin, Covid-19-The Search for Effective Therapy, N Engl J Med, doi:10.1056/NEJMe2005477
Bansal, Zheng, Song, The role of the active site flap in streptavidin/biotin complex formation, J Am Chem Soc, doi:10.1021/jacs.8b00743.
Barretto, Jukneliene, Ratia, The papainlike protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity, J Virol, doi:10.1128/JVI.79.24.15189-15198.2005
Berman, Westbrook, Feng, The Protein Data Bank, Nucleic Acids Res, doi:10.1093/nar/28.1.235
Bhimraj, Morgan, Shumaker, Infectious diseases Society of America guidelines on the treatment and management of patients with COVID-19, Clin Infect Dis, doi:10.1093/cid/ciaa478
Brogi, Calderone, Off-target ACE2 ligands: possible therapeutic option for CoVid-19?, Br J Clin Pharmacol, doi:10.1111/bcp.14343
Cao, Wang, Wen, A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19, N Engl J Med, doi:10.1056/NEJMoa2001282
Ch Ávez Thielemann, Cardellini, Fasano, From GROMACS to LAMMPS: GRO2LAM: A converter for molecular dynamics software, J Mol Model, doi:10.1007/s00894-019-4011-x
Chan, Yao, Yeung, Treatment with Lopinavir/Ritonavir or Interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset, J Infect Dis, doi:10.1093/infdis/jiv392
Chang, Tung, Lee, Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking, Preprints, doi:10.20944/preprints202002.0242.v2
Childers, Daggett, Validating molecular dynamics simulations against experimental observables in light of underlying conformational ensembles, J Phys Chem B, doi:10.1021/acs.jpcb.8b02144
Chinnasamy, Selvaraj, Selvaraj, Combining in silico and in vitro approaches to identification of potent inhibitor against phospholipase A2 (PLA2), Int J Biol Macromol, doi:10.1016/j.ijbiomac.2019.12.091
Chu, Cheng, Hung, Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings, Thorax, doi:10.1136/thorax.2003.012658
Daczkowski, Dzimianski, Clasman, Structural Insights into the Interaction of Coronavirus Papain-Like Proteases and Interferon-Stimulated Gene Product 15 from Different Species, J Mol Biol, doi:10.1016/j.jmb.2017.04.011
Dey, Borkotoky, Banerjee, In silico identification of Tretinoin as a SARS-CoV-2 Envelope (E) protein ion channel inhibitor, Comput Biol Med, doi:10.1016/j.compbiomed.2020.104063
Friesner, Banks, Murphy, Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J Med Chem, doi:10.1021/jm0306430
Gao, Tian, Yang, Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends, doi:10.5582/bst.2020.01047
Guo, Cao, Hong, The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak -an update on the status, Mil Med Res, doi:10.1186/s40779-020-00240-0
Gupta, Zhou, Profiling SARS-CoV-2 main protease (MPRO) binding to repurposed drugs using molecular dynamics simulations in classical and neural networktrained force fields, ACS Combinatorial Science, doi:10.1021/acscombsci.0c00140
Huang, Wang, Li, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet North Am Ed, doi:10.1016/S0140-6736(20)30183-5.
Jin, Du, Xu, Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kar, Potential Anti-SARS-CoV-2 therapeutics that target the post-entry stages of the viral life cycle: A comprehensive review, Viruses, doi:10.3390/v12101092.
Khater, Das, Repurposing Ivermectin to inhibit the activity of SARS CoV2 helicase: possible implications for COVID 19 therapeutics, OSF Preprints, doi:10.31219/osf.io/8dseq
Kim, Won, Kee, Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome, Antivir Ther, doi:10.3851/IMP3002
Kouznetsova, Zhang, Tatineni, Potential COVID-19 papain-like protease PLpro inhibitors: repurposing FDAapproved drugs, Peer J, doi:10.7717/peerj.9965
Lamb, Remdesivir: first approval, Drugs, doi:10.1007/s40265-020-01378-w
Lin, Shen, He, Molecular Modeling Evaluation of the Binding Effect of Ritonavir, Lopinavir and Darunavir to Severe Acute Respiratory Syndrome Coronavirus 2 Proteases, bioRxiv, doi:10.1101/2020.01.31.929695
Liu, Grimm, Dai, CB-Dock: a web server for cavity detection-guided protein-ligand blind docking, Acta Pharmacol Sin, doi:10.1038/s41401-019-0228-6
Liu, Jiang, Liu, Computational Evaluation of the COVID-19 3c-like Protease Inhibition Mechanism, and Drug Repurposing Screening, ChemRxiv, doi:10.26434/chemrxiv.12090426
Liu, Wang, Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines, bioRxiv, doi:10.1016/j.jgg.2020.02.001
Liu, Zheng, Wang, Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus, Bioinformatics, doi:10.1093/bioinformatics/btaa224
Martin, Homology models of the papain-like protease PLpro from Coronavirus 2019-nCoV, ChemRxiv, doi:10.26434/chemrxiv.11799705.v1
Martiniano, Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach, J Mol Graph Model, doi:10.1016/j.jmgm.2020.107762
Matthay, Aldrich, Gotts, Treatment for severe acute respiratory distress syndrome from COVID-19, Lancet Respir Med, doi:10.1016/S2213-2600(20)30127-2
Mcconkey, Sobolev, Edelman, The performance of current methods in ligand-protein docking, Curr Sci
Monteleone, Sarzi-Puttini, Ardizzone, Preventing COVID-19-induced pneumonia with anticytokine therapy, Lancet Rheumatol, doi:10.1016/S2665-9913(20)30092-8
Morris, Huey, Lindstrom, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J Comput Chem, doi:10.1002/jcc.21256
Nguyen, Nguyen, Truong, Remdesivir Strongly Binds to both RNA-dependent RNA Polymerase and Main Protease of SARS-CoV-2: Evidence from Molecular Simulations, J. Phys. Chem. B, doi:10.1021/acs.jpcb.0c07312
Nukoolkarn, Lee, Malaisree, Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CLpro inhibitors, J Theor Biol, doi:10.1016/j.jtbi.2008.07.030
Nutho, Mahalapbutr, Hengphasatporn, Why are lopinavir and ritonavir effective against the newly emerged Coronavirus 2019? Atomistic insights into the inhibitory mechanisms, Biochemistry, doi:10.1021/acs.biochem.0c00160
O'boyle, Banck, James, Open Babel: An open chemical toolbox, J Cheminform, doi:10.1186/1758-2946-3-33
Oostenbrink, Villa, Mark, A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6, J Comput Chem, doi:10.1002/jcc.20090
Prajapat, Shekhar, Sarma, Virtual screening and molecular dynamics study of approved drugs as inhibitors of spike protein S1 domain and ACE2 interaction in SARS-CoV-2, J Mol Graph Model, doi:10.1016/j.jmgm.2020.107716
Rakhshani, Dehghanian, Rahati, Enhanced GROMACS: toward a better numerical simulation framework, J Mol Model, doi:10.1007/s00894-019-4232-z
Ramírez, Caballero, Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data?, Molecules, doi:10.3390/molecules23051038
Refaey, El-Ashrey, Nissan, Repurposing of renin inhibitors as SARS-COV-2 main protease inhibitors: A computational study, Virology, doi:10.1016/j.virol.2020.12.008
Reiner, Hatamipour, Banach, Statins and the COVID-19 main protease, Arch Med Sci, doi:10.5114/aoms.2020.94655
Rimanshee, Amit, Vishal, Potential inhibitors against papain-like protease of novel coronavirus(SARS-CoV-2)from FDA approved drugs, doi:10.26434/chemrxiv.11860011.v2
Rochwerg, Agarwal, Zeng, Remdesivir for severe covid-19: a clinical practice guideline, BMJ, doi:10.1136/bmj.m2924
Rodriguez-Morales, Bonilla-Aldana, Tiwari, COVID-19, an emerging coronavirus infection: current scenario and recent developments-an overview, J Pure Appl Microbiol, doi:10.22207/JPAM.14.1.02
Rothe, Schunk, Sothmann, Transmission of 2019-nCoV infection from an asymptomatic contact in Germany, N Engl J Med, doi:10.1056/NEJMc2001468
Ruiz-Carmona, Alvarez-Garcia, Foloppe, rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids, PLoS Comput Biol, doi:10.1371/journal.pcbi.1003571
Snijder, Bredenbeek, Dobbe, Unique and conserved features of genome and proteome of SARScoronavirus, an early split-off from the coronavirus group 2 lineage, J Mol Biol, doi:10.1016/S0022-2836(03)00865-9
Sterling, Irwin, ZINC 15-Ligand discovery for everyone, J Chem Inf Model, doi:10.1021/acs.jcim.5b00559
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem, doi:10.1002/jcc.21334
Verity, Okell, Dorigatti, Estimates of the severity of coronavirus disease 2019: a model-based analysis, Lancet Infect Dis, doi:10.1016/S1473-3099(20)30243-7
Wang, Cao, Zhang, Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res, doi:10.1038/s41422-020-0282-0
Wrapp, Wang, Corbett, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, doi:10.1126/science.abb2507
Wu, Liu, Yang, Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharm Sin B, doi:10.1016/j.apsb.2020.02.008
Wu, Zhao, Yu, A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3.
Xu, Peng, Shi, Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation, bioRxiv, doi:10.1101/2020.01.27.92162
Yin, Mao, Luan, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science, doi:10.1126/science.abc1560
Yin, Mao, Luan, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science, doi:10.1126/science.abc1560
Ziebuhr, Siddell, Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab, J Virol, doi:10.1128/JVI.73.1.177-185.1999
DOI record:
{
"DOI": "10.1093/pcmedi/pbab001",
"ISSN": [
"2096-5303",
"2516-1571"
],
"URL": "http://dx.doi.org/10.1093/pcmedi/pbab001",
"abstract": "<jats:title>Abstract</jats:title><jats:p>The pandemic of novel coronavirus disease 2019 (COVID-19) has rampaged the world, with more than 58.4 million confirmed cases and over 1.38 million deaths across the world by 23 November 2020. There is an urgent need to identify effective drugs and vaccines to fight against the virus. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the family of coronaviruses consisting of four structural and 16 non-structural proteins (NSP). Three non-structural proteins, main protease (Mpro), papain-like protease (PLpro), and RNA-dependent RNA polymerase (RdRp), are believed to have a crucial role in replication of the virus. We applied computational ligand-receptor binding modeling and performed comprehensive virtual screening on FDA-approved drugs against these three SARS-CoV-2 proteins using AutoDock Vina, Glide, and rDock. Our computational studies identified six novel ligands as potential inhibitors against SARS-CoV-2, including antiemetics rolapitant and ondansetron for Mpro; labetalol and levomefolic acid for PLpro; and leucal and antifungal natamycin for RdRp. Molecular dynamics simulation confirmed the stability of the ligand-protein complexes. The results of our analysis with some other suggested drugs indicated that chloroquine and hydroxychloroquine had high binding energy (low inhibitory effect) with all three proteins—Mpro, PLpro, and RdRp. In summary, our computational molecular docking approach and virtual screening identified some promising candidate SARS-CoV-2 inhibitors that may be considered for further clinical studies.</jats:p>",
"author": [
{
"affiliation": [
{
"name": "Center for Genomics, School of Medicine, Loma Linda University, Loma Linda, CA 92350, USA"
}
],
"family": "Hosseini",
"given": "Maryam",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Center for Genomics, School of Medicine, Loma Linda University, Loma Linda, CA 92350, USA"
}
],
"family": "Chen",
"given": "Wanqiu",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0147-2758",
"affiliation": [
{
"name": "Lawrence D. Longo, MD Center for Perinatal Biology, Department of Basic Sciences, School of Medicine, Loma Linda University, Loma Linda, CA 92350, USA"
}
],
"authenticated-orcid": false,
"family": "Xiao",
"given": "Daliao",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8861-2121",
"affiliation": [
{
"name": "Center for Genomics, School of Medicine, Loma Linda University, Loma Linda, CA 92350, USA"
},
{
"name": "Division of Microbiology & Molecular Genetics, Department of Basic Sciences, School of Medicine, Loma Linda University, Loma Linda, CA 92350, USA"
}
],
"authenticated-orcid": false,
"family": "Wang",
"given": "Charles",
"sequence": "additional"
}
],
"container-title": "Precision Clinical Medicine",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
1,
18
]
],
"date-time": "2021-01-18T20:04:27Z",
"timestamp": 1611000267000
},
"deposited": {
"date-parts": [
[
2022,
12,
12
]
],
"date-time": "2022-12-12T02:16:32Z",
"timestamp": 1670811392000
},
"funder": [
{
"DOI": "10.13039/100000968",
"award": [
"18IPA34170301"
],
"doi-asserted-by": "publisher",
"name": "American Heart Association"
},
{
"DOI": "10.13039/100000002",
"award": [
"R01/HD088039"
],
"doi-asserted-by": "publisher",
"name": "National Institutes of Health"
}
],
"indexed": {
"date-parts": [
[
2023,
3,
27
]
],
"date-time": "2023-03-27T23:30:59Z",
"timestamp": 1679959859909
},
"is-referenced-by-count": 41,
"issue": "1",
"issued": {
"date-parts": [
[
2021,
1,
18
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2021,
1,
18
]
]
},
"published-print": {
"date-parts": [
[
2021,
4,
3
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
1,
18
]
],
"date-time": "2021-01-18T00:00:00Z",
"timestamp": 1610928000000
}
}
],
"link": [
{
"URL": "http://academic.oup.com/pcm/advance-article-pdf/doi/10.1093/pcmedi/pbab001/36623673/pbab001.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "http://academic.oup.com/pcm/article-pdf/4/1/1/36872109/pbab001.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "syndication"
},
{
"URL": "http://academic.oup.com/pcm/article-pdf/4/1/1/36872109/pbab001.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"page": "1-16",
"prefix": "10.1093",
"published": {
"date-parts": [
[
2021,
1,
18
]
]
},
"published-online": {
"date-parts": [
[
2021,
1,
18
]
]
},
"published-other": {
"date-parts": [
[
2021,
3
]
]
},
"published-print": {
"date-parts": [
[
2021,
4,
3
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference": [
{
"DOI": "10.1038/s41586-020-2008-3.",
"article-title": "A new coronavirus associated with human respiratory disease in China",
"author": "Wu",
"doi-asserted-by": "publisher",
"first-page": "265",
"journal-title": "Nature",
"key": "2021040614470645800_bib1",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5.",
"article-title": "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China",
"author": "Huang",
"doi-asserted-by": "publisher",
"first-page": "497",
"journal-title": "Lancet North Am Ed",
"key": "2021040614470645800_bib2",
"volume": "395",
"year": "2020"
},
{
"key": "2021040614470645800_bib3",
"year": "2020"
},
{
"DOI": "10.22207/JPAM.14.1.02",
"article-title": "COVID-19, an emerging coronavirus infection: current scenario and recent developments-an overview",
"author": "Rodriguez-Morales",
"doi-asserted-by": "publisher",
"first-page": "6150",
"journal-title": "J Pure Appl Microbiol",
"key": "2021040614470645800_bib4",
"volume": "14",
"year": "2020"
},
{
"key": "2021040614470645800_bib5",
"year": "2003"
},
{
"key": "2021040614470645800_bib6",
"year": "2019"
},
{
"DOI": "10.1056/NEJMc2001468",
"article-title": "Transmission of 2019-nCoV infection from an asymptomatic contact in Germany",
"author": "Rothe",
"doi-asserted-by": "publisher",
"first-page": "970",
"journal-title": "N Engl J Med",
"key": "2021040614470645800_bib7",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/S1473-3099(20)30243-7",
"article-title": "Estimates of the severity of coronavirus disease 2019: a model-based analysis",
"author": "Verity",
"doi-asserted-by": "publisher",
"first-page": "669",
"journal-title": "Lancet Infect Dis",
"key": "2021040614470645800_bib8",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1056/NEJMe2005477",
"article-title": "Covid-19—The Search for Effective Therapy",
"author": "Baden",
"doi-asserted-by": "publisher",
"first-page": "1851",
"journal-title": "N Engl J Med",
"key": "2021040614470645800_bib9",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/S2665-9913(20)30092-8",
"article-title": "Preventing COVID-19-induced pneumonia with anticytokine therapy",
"author": "Monteleone",
"doi-asserted-by": "publisher",
"first-page": "255",
"issue": "5",
"journal-title": "Lancet Rheumatol",
"key": "2021040614470645800_bib10",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1016/S2213-2600(20)30127-2",
"article-title": "Treatment for severe acute respiratory distress syndrome from COVID-19",
"author": "Matthay",
"doi-asserted-by": "publisher",
"first-page": "433",
"issue": "5",
"journal-title": "Lancet Respir Med",
"key": "2021040614470645800_bib11",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1016/j.apsb.2020.02.008",
"article-title": "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods",
"author": "Wu",
"doi-asserted-by": "publisher",
"first-page": "766",
"issue": "5",
"journal-title": "Acta Pharm Sin B",
"key": "2021040614470645800_bib12",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1186/s40779-020-00240-0",
"article-title": "The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status",
"author": "Guo",
"doi-asserted-by": "publisher",
"first-page": "11",
"journal-title": "Mil Med Res",
"key": "2021040614470645800_bib13",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1128/JVI.73.1.177-185.1999",
"article-title": "Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab",
"author": "Ziebuhr",
"doi-asserted-by": "publisher",
"first-page": "177",
"journal-title": "J Virol",
"key": "2021040614470645800_bib14",
"volume": "73",
"year": "1999"
},
{
"DOI": "10.1128/JVI.79.24.15189-15198.2005",
"article-title": "The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity",
"author": "Barretto",
"doi-asserted-by": "publisher",
"first-page": "15189",
"journal-title": "J Virol",
"key": "2021040614470645800_bib15",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1016/S0022-2836(03)00865-9",
"article-title": "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage",
"author": "Snijder",
"doi-asserted-by": "publisher",
"first-page": "991",
"journal-title": "J Mol Biol",
"key": "2021040614470645800_bib16",
"volume": "331",
"year": "2003"
},
{
"DOI": "10.1093/cid/ciaa478",
"article-title": "Infectious diseases Society of America guidelines on the treatment and management of patients with COVID-19",
"author": "Bhimraj",
"doi-asserted-by": "publisher",
"journal-title": "Clin Infect Dis",
"key": "2021040614470645800_bib17",
"year": "2020"
},
{
"DOI": "10.1021/acs.biochem.0c00160",
"article-title": "Why are lopinavir and ritonavir effective against the newly emerged Coronavirus 2019? Atomistic insights into the inhibitory mechanisms",
"author": "Nutho",
"doi-asserted-by": "publisher",
"first-page": "1769",
"journal-title": "Biochemistry",
"key": "2021040614470645800_bib18",
"volume": "59",
"year": "2020"
},
{
"key": "2021040614470645800_bib19",
"year": "2020"
},
{
"DOI": "10.1126/science.abc1560",
"article-title": "Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir",
"author": "Yin",
"doi-asserted-by": "publisher",
"first-page": "1499",
"issue": "6498",
"journal-title": "Science",
"key": "2021040614470645800_bib20",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1136/thorax.2003.012658",
"article-title": "Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings",
"author": "Chu",
"doi-asserted-by": "publisher",
"first-page": "252",
"journal-title": "Thorax",
"key": "2021040614470645800_bib21",
"volume": "59",
"year": "2004"
},
{
"DOI": "10.1093/infdis/jiv392",
"article-title": "Treatment with Lopinavir/Ritonavir or Interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset",
"author": "Chan",
"doi-asserted-by": "publisher",
"first-page": "1904",
"issue": "12",
"journal-title": "J Infect Dis",
"key": "2021040614470645800_bib22",
"volume": "212",
"year": "2015"
},
{
"article-title": "Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome",
"author": "Kim",
"first-page": "455",
"journal-title": "Antivir Ther",
"key": "2021040614470645800_bib23",
"volume": "21",
"year": "2016"
},
{
"DOI": "10.1056/NEJMoa2001282",
"article-title": "A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19",
"author": "Cao",
"doi-asserted-by": "publisher",
"first-page": "1787",
"journal-title": "N Engl J Med",
"key": "2021040614470645800_bib24",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "269",
"journal-title": "Cell Res",
"key": "2021040614470645800_bib25",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.5582/bst.2020.01047",
"article-title": "Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies",
"author": "Gao",
"doi-asserted-by": "publisher",
"first-page": "72",
"journal-title": "Biosci Trends",
"key": "2021040614470645800_bib26",
"volume": "14",
"year": "2020"
},
{
"DOI": "10.26434/chemrxiv.11860011.v2",
"article-title": "Potential inhibitors against papain-like protease of novel coronavirus(SARS-CoV-2)from FDA approved drugs",
"author": "Rimanshee",
"doi-asserted-by": "publisher",
"key": "2021040614470645800_bib27",
"year": "2020"
},
{
"DOI": "10.1101/2020.01.27.92162",
"article-title": "Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation",
"author": "Xu",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv",
"key": "2021040614470645800_bib28",
"year": "2020"
},
{
"DOI": "10.1016/j.jgg.2020.02.001",
"article-title": "Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines",
"author": "Liu",
"doi-asserted-by": "publisher",
"first-page": "119",
"journal-title": "bioRxiv",
"key": "2021040614470645800_bib29",
"volume": "47",
"year": "2020"
},
{
"DOI": "10.20944/preprints202002.0242.v2",
"article-title": "Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking",
"author": "Chang",
"doi-asserted-by": "publisher",
"key": "2021040614470645800_bib30",
"volume-title": "Preprints",
"year": "2020"
},
{
"article-title": "Molecular Modeling Evaluation of the Binding Effect of Ritonavir, Lopinavir and Darunavir to Severe Acute Respiratory Syndrome Coronavirus 2 Proteases",
"author": "Lin",
"journal-title": "bioRxiv",
"key": "2021040614470645800_bib31",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors",
"author": "Jin",
"doi-asserted-by": "publisher",
"first-page": "289",
"key": "2021040614470645800_bib32",
"volume-title": "Nature",
"year": "2020"
},
{
"DOI": "10.1093/nar/28.1.235",
"article-title": "The Protein Data Bank",
"author": "Berman",
"doi-asserted-by": "crossref",
"first-page": "235",
"journal-title": "Nucleic Acids Res",
"key": "2021040614470645800_bib33",
"volume": "28",
"year": "2000"
},
{
"DOI": "10.1016/j.jmb.2017.04.011",
"article-title": "Structural Insights into the Interaction of Coronavirus Papain-Like Proteases and Interferon-Stimulated Gene Product 15 from Different Species",
"author": "Daczkowski",
"doi-asserted-by": "publisher",
"first-page": "1661",
"journal-title": "J Mol Biol",
"key": "2021040614470645800_bib34",
"volume": "429",
"year": "2017"
},
{
"DOI": "10.26434/chemrxiv.11799705.v1",
"article-title": "Homology models of the papain-like protease PLpro from Coronavirus 2019-nCoV",
"author": "Martin",
"doi-asserted-by": "publisher",
"key": "2021040614470645800_bib35",
"volume-title": "ChemRxiv",
"year": "2020"
},
{
"DOI": "10.1021/acs.jcim.5b00559",
"article-title": "ZINC 15–Ligand discovery for everyone",
"author": "Sterling",
"doi-asserted-by": "publisher",
"first-page": "2324",
"journal-title": "J Chem Inf Model",
"key": "2021040614470645800_bib36",
"volume": "55",
"year": "2015"
},
{
"author": "Schrödinger LigPrep",
"key": "2021040614470645800_bib37"
},
{
"DOI": "10.1002/jcc.21256",
"article-title": "AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility",
"author": "Morris",
"doi-asserted-by": "publisher",
"first-page": "2785",
"journal-title": "J Comput Chem",
"key": "2021040614470645800_bib38",
"volume": "30",
"year": "2009"
},
{
"DOI": "10.1186/1758-2946-3-33",
"article-title": "Open Babel: An open chemical toolbox",
"author": "O'Boyle",
"doi-asserted-by": "publisher",
"first-page": "33",
"journal-title": "J Cheminform",
"key": "2021040614470645800_bib39",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.1007/s00894-019-4011-x",
"article-title": "From GROMACS to LAMMPS: GRO2LAM: A converter for molecular dynamics software",
"author": "Chávez Thielemann",
"doi-asserted-by": "publisher",
"first-page": "147",
"journal-title": "J Mol Model",
"key": "2021040614470645800_bib40",
"volume": "25",
"year": "2019"
},
{
"DOI": "10.1007/s00894-019-4232-z",
"article-title": "Enhanced GROMACS: toward a better numerical simulation framework",
"author": "Rakhshani",
"doi-asserted-by": "publisher",
"first-page": "355",
"journal-title": "J Mol Model",
"key": "2021040614470645800_bib41",
"volume": "25",
"year": "2019"
},
{
"DOI": "10.1002/jcc.20090",
"article-title": "A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6",
"author": "Oostenbrink",
"doi-asserted-by": "publisher",
"first-page": "1656",
"journal-title": "J Comput Chem",
"key": "2021040614470645800_bib42",
"volume": "25",
"year": "2004"
},
{
"DOI": "10.1021/acs.jpcb.8b02144",
"article-title": "Validating molecular dynamics simulations against experimental observables in light of underlying conformational ensembles",
"author": "Childers",
"doi-asserted-by": "publisher",
"first-page": "6673",
"journal-title": "J Phys Chem B",
"key": "2021040614470645800_bib43",
"volume": "122",
"year": "2018"
},
{
"DOI": "10.1016/j.ijbiomac.2019.12.091",
"article-title": "Combining in silico and in vitro approaches to identification of potent inhibitor against phospholipase A2 (PLA2)",
"author": "Chinnasamy",
"doi-asserted-by": "publisher",
"first-page": "53",
"journal-title": "Int J Biol Macromol",
"key": "2021040614470645800_bib44",
"volume": "144",
"year": "2020"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "publisher",
"first-page": "455",
"journal-title": "J Comput Chem",
"key": "2021040614470645800_bib45",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1021/jm0306430",
"article-title": "Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy",
"author": "Friesner",
"doi-asserted-by": "publisher",
"first-page": "1739",
"journal-title": "J Med Chem",
"key": "2021040614470645800_bib46",
"volume": "47",
"year": "2004"
},
{
"DOI": "10.1371/journal.pcbi.1003571",
"article-title": "rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids",
"author": "Ruiz-Carmona",
"doi-asserted-by": "publisher",
"first-page": "e1003571",
"journal-title": "PLoS Comput Biol",
"key": "2021040614470645800_bib47",
"volume": "10",
"year": "2014"
},
{
"DOI": "10.3390/molecules23051038",
"article-title": "Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data?",
"author": "Ramírez",
"doi-asserted-by": "publisher",
"journal-title": "Molecules",
"key": "2021040614470645800_bib48",
"volume": "23",
"year": "2018"
},
{
"DOI": "10.1016/j.arabjc.2020.08.004",
"article-title": "Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics",
"author": "Alamri",
"doi-asserted-by": "publisher",
"first-page": "7224",
"journal-title": "Arabian J Chem",
"key": "2021040614470645800_bib49",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1126/science.abc1560",
"article-title": "Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir",
"author": "Yin",
"doi-asserted-by": "publisher",
"first-page": "1499",
"journal-title": "Science",
"key": "2021040614470645800_bib50",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.5114/aoms.2020.94655",
"article-title": "Statins and the COVID-19 main protease",
"author": "Reiner",
"doi-asserted-by": "publisher",
"first-page": "490",
"journal-title": "Arch Med Sci",
"key": "2021040614470645800_bib51",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1093/bioinformatics/btaa224",
"article-title": "Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus",
"author": "Liu",
"doi-asserted-by": "publisher",
"first-page": "3295",
"journal-title": "Bioinformatics",
"key": "2021040614470645800_bib52",
"volume": "36",
"year": "2020"
},
{
"DOI": "10.1021/acs.jpcb.0c07312",
"article-title": "Remdesivir Strongly Binds to both RNA-dependent RNA Polymerase and Main Protease of SARS-CoV-2: Evidence from Molecular Simulations",
"author": "Nguyen",
"doi-asserted-by": "publisher",
"first-page": "11337",
"key": "2021040614470645800_bib53",
"volume-title": "J. Phys. Chem. B",
"year": "2020"
},
{
"DOI": "10.3390/v12101092.",
"article-title": "Potential Anti-SARS-CoV-2 therapeutics that target the post-entry stages of the viral life cycle: A comprehensive review",
"author": "Al-Horani",
"doi-asserted-by": "publisher",
"first-page": "1092",
"journal-title": "Viruses",
"key": "2021040614470645800_bib54",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1007/s40265-020-01378-w",
"article-title": "Remdesivir: first approval",
"author": "Lamb",
"doi-asserted-by": "publisher",
"first-page": "1355",
"journal-title": "Drugs",
"key": "2021040614470645800_bib55",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1136/bmj.m2924",
"article-title": "Remdesivir for severe covid-19: a clinical practice guideline",
"author": "Rochwerg",
"doi-asserted-by": "publisher",
"first-page": "m2924",
"journal-title": "BMJ",
"key": "2021040614470645800_bib56",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1016/j.jtbi.2008.07.030",
"article-title": "Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CLpro inhibitors",
"author": "Nukoolkarn",
"doi-asserted-by": "publisher",
"first-page": "861",
"journal-title": "J Theor Biol",
"key": "2021040614470645800_bib57",
"volume": "254",
"year": "2008"
},
{
"article-title": "The performance of current methods in ligand–protein docking",
"author": "McConkey",
"first-page": "845",
"journal-title": "Curr Sci",
"key": "2021040614470645800_bib58",
"volume": "83",
"year": "2002"
},
{
"DOI": "10.1021/jacs.8b00743.",
"article-title": "The role of the active site flap in streptavidin/biotin complex formation",
"author": "Bansal",
"doi-asserted-by": "publisher",
"first-page": "5434",
"journal-title": "J Am Chem Soc",
"key": "2021040614470645800_bib59",
"volume": "140",
"year": "2018"
},
{
"DOI": "10.1038/s41401-019-0228-6",
"article-title": "CB-Dock: a web server for cavity detection-guided protein–ligand blind docking",
"author": "Liu",
"doi-asserted-by": "publisher",
"first-page": "138",
"journal-title": "Acta Pharmacol Sin",
"key": "2021040614470645800_bib60",
"volume": "41",
"year": "2020"
},
{
"DOI": "10.26434/chemrxiv.12090426",
"article-title": "Computational Evaluation of the COVID-19 3c-like Protease Inhibition Mechanism, and Drug Repurposing Screening",
"author": "Liu",
"doi-asserted-by": "publisher",
"key": "2021040614470645800_bib61",
"volume-title": "ChemRxiv",
"year": "2020"
},
{
"DOI": "10.1016/j.virol.2020.12.008",
"article-title": "Repurposing of renin inhibitors as SARS-COV-2 main protease inhibitors: A computational study",
"author": "",
"doi-asserted-by": "publisher",
"first-page": "48",
"key": "2021040614470645800_bib62",
"volume-title": "Virology",
"year": "2021"
},
{
"DOI": "10.1016/j.compbiomed.2020.104063",
"article-title": "In silico identification of Tretinoin as a SARS-CoV-2 Envelope (E) protein ion channel inhibitor",
"author": "Dey",
"doi-asserted-by": "publisher",
"first-page": "104063",
"journal-title": "Comput Biol Med",
"key": "2021040614470645800_bib63",
"volume": "127",
"year": "2020"
},
{
"DOI": "10.1016/j.jmgm.2020.107762",
"article-title": "Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach",
"author": "Martiniano",
"doi-asserted-by": "publisher",
"first-page": "107762",
"journal-title": "J Mol Graph Model",
"key": "2021040614470645800_bib64",
"volume": "101",
"year": "2020"
},
{
"DOI": "10.1016/j.jmgm.2020.107716",
"article-title": "Virtual screening and molecular dynamics study of approved drugs as inhibitors of spike protein S1 domain and ACE2 interaction in SARS-CoV-2",
"author": "Prajapat",
"doi-asserted-by": "publisher",
"first-page": "107716",
"journal-title": "J Mol Graph Model",
"key": "2021040614470645800_bib65",
"volume": "101",
"year": "2020"
},
{
"DOI": "10.7717/peerj.9965",
"article-title": "Potential COVID-19 papain-like protease PLpro inhibitors: repurposing FDA-approved drugs",
"author": "Kouznetsova",
"doi-asserted-by": "publisher",
"first-page": "e9965",
"journal-title": "Peer J",
"key": "2021040614470645800_bib66",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1126/science.abb2507",
"article-title": "Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation",
"author": "Wrapp",
"doi-asserted-by": "publisher",
"first-page": "1260",
"journal-title": "Science",
"key": "2021040614470645800_bib67",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1111/bcp.14343",
"article-title": "Off-target ACE2 ligands: possible therapeutic option for CoVid-19?",
"author": "Brogi",
"doi-asserted-by": "publisher",
"first-page": "1178",
"journal-title": "Br J Clin Pharmacol",
"key": "2021040614470645800_bib68",
"volume": "86",
"year": "2020"
},
{
"article-title": "Drug repurposing to identify therapeutics against COVID 19 with SARS-Cov-2 spike glycoprotein and main protease as targets: an in silico study",
"author": "Arun",
"key": "2021040614470645800_bib69",
"volume-title": "ChemRxiv",
"year": "2020"
},
{
"DOI": "10.12688/f1000research.26359.1",
"article-title": "Targeting SARS-CoV-2 RNA-dependent RNA polymerase: An in silico drug repurposing for COVID-19",
"author": "Baby",
"doi-asserted-by": "publisher",
"first-page": "1166",
"journal-title": "F1000Research",
"key": "2021040614470645800_bib70",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1021/acscombsci.0c00140",
"article-title": "Profiling SARS-CoV-2 main protease (MPRO) binding to repurposed drugs using molecular dynamics simulations in classical and neural network-trained force fields",
"author": "Gupta",
"doi-asserted-by": "publisher",
"first-page": "826",
"key": "2021040614470645800_bib71",
"volume-title": "ACS Combinatorial Science",
"year": "2020"
},
{
"DOI": "10.31219/osf.io/8dseq",
"article-title": "Repurposing Ivermectin to inhibit the activity of SARS CoV2 helicase: possible implications for COVID 19 therapeutics",
"author": "Khater",
"doi-asserted-by": "crossref",
"key": "2021040614470645800_bib72",
"volume-title": "OSF Preprints",
"year": "2020"
},
{
"DOI": "10.1038/s41598-020-75762-7",
"article-title": "Searching for target-specific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach",
"author": "Arul",
"doi-asserted-by": "publisher",
"first-page": "19125",
"key": "2021040614470645800_bib73",
"volume-title": "Sci Rep",
"year": "2020"
}
],
"reference-count": 73,
"references-count": 73,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/pcm/article/4/1/1/6103931"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Medicine"
],
"subtitle": [],
"title": "Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs",
"type": "journal-article",
"volume": "4"
}