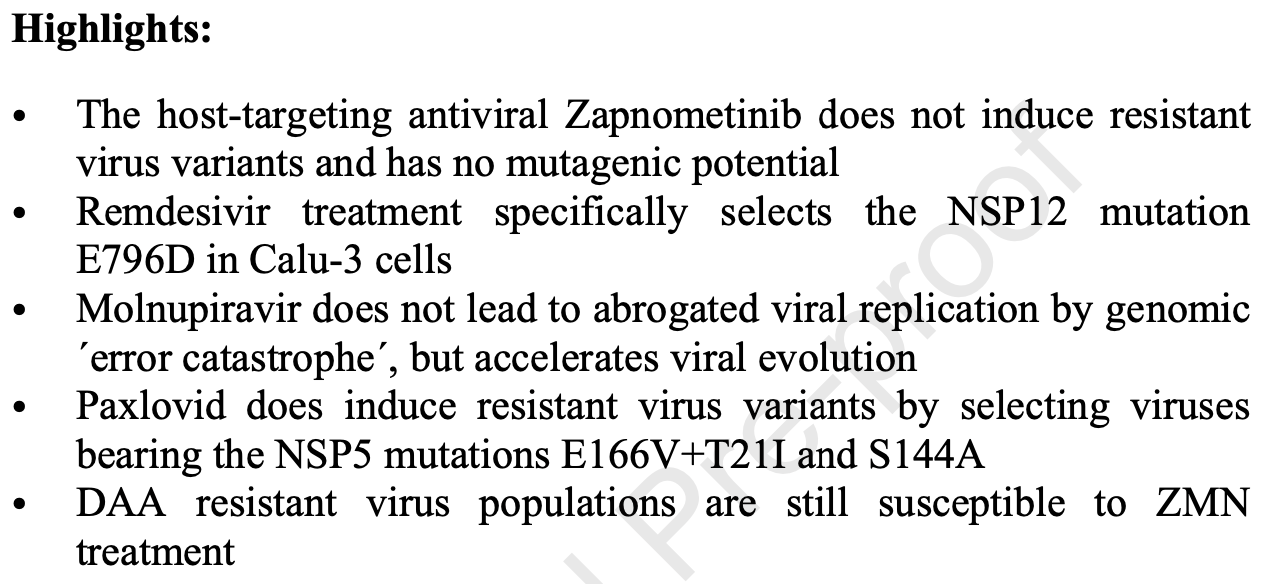
The host-targeted antiviral drug Zapnometinib exhibits a high barrier to the development of SARS-CoV-2 resistance
et al., Antiviral Research, doi:10.1016/j.antiviral.2024.105840, Mar 2024
In vitro study showing that molnupiravir and paxlovid induced resistant variants in SARS-CoV-2 during serial passaging, while the host-directed antiviral zapnometinib did not.
Authors found that molnupiravir did not lead to abrogated viral replication by genomic "error catastrophe" as postulated, but rather accelerated viral evolution leading to reduced drug susceptibility. Paxlovid treatment specifically selected viruses bearing the NSP5 mutations E166V+T21I and S144A. In contrast, zapnometinib treatment did not induce any specific mutations or reduced drug susceptibility even after 30 passages in Calu-3 and CaCo2 cells.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
Study covers molnupiravir and paxlovid.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Schreiber et al., 2 Mar 2024, peer-reviewed, 8 authors.
Contact: ludwigs@uni-muenster.de.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
The host-targeted antiviral drug Zapnometinib exhibits a high barrier to the development of SARS-CoV-2 resistance
Antiviral Research, doi:10.1016/j.antiviral.2024.105840
Host targeting antiviral drugs (HTA) are directed against cellular mechanisms which can be exploited by viruses. These mechanisms are essential for viral replication, because missing functions cannot be compensated by the virus. However, this assumption needs experimental proof. Here we compared the HTA Zapnometinib (ZMN), with direct acting antivirals (DAA) (Remdesivir (RDV), Molnupiravir (MPV), Nirmatrelvir (NTV), Ritonavir (RTV), Paxlovid PAX)), in terms of their potency to induce reduced drug susceptibilities in SARS-CoV-2. During serial passage of δ-B1.617.2 adaptation to all DAAs occurred, while the inhibitory capacity of ZMN was not altered. Known single nucleotide polymorphisms (SNPs) responsible for partial resistances were found for RDV, NTV and PAX. Additionally, the high mutagenic potential of MPV was confirmed and decreased drug efficacies were found for the first time. Reduced DAA efficacy did not alter the inhibitory potential of ZMN. These results show that ZMN confers a high barrier towards the development of viral resistance and has the potential to act against partially DAA-insensitive viruses.
Ethics statement This study does not reveal new information about the creation of resistant SARS-CoV-2 variants, nor about the resistant introducing potential of SNPs against licensed anti-SARS-CoV-2 drugs. All methods used in this study and all mutations found to induce RDV and NTV / PAX reduced susceptibilities were described before. The malicious release of these resistant virus variants would not pose a high risk, as these viruses are not fully but partially resistant and do not possess cross-resistances to other antiviral drugs. Lastly, resistant viruses did not show a replication benefit in a drug-free environment. The necessity and safety of the J o u r n a l P r e -p r o o f conducted experiments was discussed and confirmed during a risk assessment meeting with the independent bio-risk consortium of the German Federal Research Institute for Animal Health (Friedrich-Loeffler-Insitute, Greifswald-Riems).
Declaration of interests ☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. ☐The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
References
Alam, Mostafa, Kanrai, Verapamil has Antiviral Activities that Target Different Steps of the Influenza Virus Replication Cycle, J. Antivir. Antiretrovir, doi:10.4172/jaa.1000147
Droebner, Pleschka, Ludwig, Planz, Antiviral activity of the MEKinhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo, Antiviral Res, doi:10.1016/j.antiviral.2011.08.002
Götte, The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance, Curr. Opin. Virol, doi:10.1016/j.coviro.2012.08.004
Hopcraft, Evans, Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins, Hepatology, doi:10.1002/hep.27815
Hu, Lewandowski, Tan, Zhang, Morgan et al., Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug J o u r n a l P r e -p r o o f Resistance to Nirmatrelvir, ACS Cent. Sci, doi:10.1021/acscentsci.3c00538
Hull, Montaner, Ritonavir-boosted protease inhibitors in HIV therapy, Annals of Medicine, doi:10.3109/07853890.2011.572905
Iketani, Mohri, Culbertson, Hong, Duan et al., Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature, doi:10.1038/s41586-022-05514-2
J O U R N A L P R E, None
J O U R N A L P R E, None
Karamitros, Papadopoulou, Bousali, Mexias, Tsiodras et al., SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies, J. Clin. Virol, doi:10.1016/j.jcv.2020.104585
Khandelwal, Chander, Rawat, Riyesh, Nishanth et al., Emetine inhibits replication of RNA and DNA viruses without generating drug-resistant virus variants, Antiviral Res, doi:10.1016/j.antiviral.2017.06.006
Krumm, Ndungu, Yoon, Dochow, Sun et al., Potent host-directed small-molecule inhibitors of myxovirus rna-dependent rna-polymerases, PLoS One, doi:10.1371/journal.pone.0020069
Kumar, Khandelwal, Chander, Riyesh, Tripathi et al., MNK1 inhibitor as an antiviral agent suppresses buffalopox virus protein synthesis, Antiviral Res, doi:10.1016/j.antiviral.2018.10.022
Kumar, Khandelwal, Kumar, Chander, Rawat et al., Inhibitor of sarco/endoplasmic reticulum calcium-ATPase impairs multiple steps of paramyxovirus replication, Front. Microbiol, doi:10.3389/fmicb.2019.00209
Kumar, Sharma, Ly, Parslow, Liang, Receptor tyrosine kinase inhibitors that block replication of influenza A and other viruses, Antimicrob. Agents Chemother, doi:10.1128/AAC.00725-11
Lauring, Andino, Quasispecies theory and the behavior of RNA viruses, PLoS Pathog, doi:10.1371/journal.ppat.1001005
Ludwig, Wolff, Ehrhardt, Wurzer, Reinhardt et al., MEK inhibition impairs influenza B virus propagation without emergence of resistant variants, FEBS Lett, doi:10.1016/S0014-5793(04)00108-5
Martinot, Jary, Fafi-Kremer, Leducq, Delagreverie et al., Emerging RNA-Dependent RNA Polymerase Mutation in a Remdesivir-Treated B-cell Immunodeficient Patient with Protracted Coronavirus Disease, Clin. Infect. Dis, doi:10.1093/cid/ciaa1474
Mccrone, Lauring, Genetic bottlenecks in intraspecies virus transmission, Curr. Opin. Virol, doi:10.1016/j.coviro.2017.10.008
Moghadasi, Heilmann, Khalil, Nnabuife, Kearns et al., Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors, Sci. Adv, doi:10.1126/sciadv.ade8778
Pleschka, Wolff, Ehrhardt, Hobom, Planz et al., Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade, Nat. Cell Biol, doi:10.1038/35060098
Preugschas, Hrincius, Mewis, Tran, Ludwig et al., Late activation of the Raf/MEK/ERK pathway is required for translocation of the respiratory syncytial virus F protein to the plasma membrane and efficient viral replication, Cell. Microbiol, doi:10.1111/cmi.12955
Puyang, Poulin, Mathy, Anderson, Ma et al., Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors, Antimicrob. Agents Chemother, doi:10.1128/AAC.01236-09
Sanderson, Hisner, Donovan-Banfield, Hartman, Lochen et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6
Sayan, Hulagu, Karatayli, Multidrug-resistant hepatitis B virus strain in a chronic Turkish patient, Hepat. Mon
Schreiber, Ambrosy, Planz, Schloer, Rescher et al., The MEK1/2 Inhibitor ATR-002 (Zapnometinib) Synergistically Potentiates the Antiviral Effect of Direct-Acting Anti-SARS-CoV-2 Drugs, Pharmaceutics, doi:10.3390/pharmaceutics14091776
Schreiber, Viemann, Schöning, Schloer, Mecate Zambrano et al., The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates proinflammatory cytokine/chemokine responses, Cell. Mol. Life Sci, doi:10.1007/s00018-021-04085-1
Shyr, Cheng, Lo, Zheng, Drug combination therapy for emerging viral diseases, Drug Discov. Today, doi:10.1016/j.drudis.2021.05.008
Takashita, Fujisaki, Yokoyama, Shirakura, Morita et al., In vitro characterization of multidrug-resistant influenza a(H1n1)pdm09 viruses carrying a dual neuraminidase mutation isolated from immunocompromised patients, Pathogens, doi:10.3390/pathogens9090725
Van Der Schaar, Van Der Linden, Lanke, Strating, Pürstinger et al., Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication, Cell Res, doi:10.1038/cr.2012.129
Wagoner, Herring, Hsiang, Ianevski, Biering et al., Combinations of host-and virus-targeting antiviral drugs confer synergistic suppression of SARS-CoV-2, Microbiol Spectr, doi:10.1128/spectrum.03331-22
Ölschläger, Pleschka, Fischer, Rziha, Wurzer et al., Lung-specific expression of active Raf kinase results in increased mortality of influenza A virus-infected mice, Oncogene, doi:10.1038/sj.onc.1207883
DOI record:
{
"DOI": "10.1016/j.antiviral.2024.105840",
"ISSN": [
"0166-3542"
],
"URL": "http://dx.doi.org/10.1016/j.antiviral.2024.105840",
"alternative-id": [
"S0166354224000482"
],
"article-number": "105840",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "The host-targeted antiviral drug Zapnometinib exhibits a high barrier to the development of SARS-CoV-2 resistance"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Antiviral Research"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.antiviral.2024.105840"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2024 The Authors. Published by Elsevier B.V."
}
],
"author": [
{
"affiliation": [],
"family": "Schreiber",
"given": "André",
"sequence": "first"
},
{
"affiliation": [],
"family": "Rodner",
"given": "Franziska",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Oberberg",
"given": "Nicole",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Anhlan",
"given": "Darisuren",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bletz",
"given": "Stefan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0649-5185",
"affiliation": [],
"authenticated-orcid": false,
"family": "Mellmann",
"given": "Alexander",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3701-943X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Planz",
"given": "Oliver",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4490-3052",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ludwig",
"given": "Stephan",
"sequence": "additional"
}
],
"container-title": "Antiviral Research",
"container-title-short": "Antiviral Research",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2024,
3,
2
]
],
"date-time": "2024-03-02T16:15:06Z",
"timestamp": 1709396106000
},
"deposited": {
"date-parts": [
[
2024,
3,
2
]
],
"date-time": "2024-03-02T19:33:13Z",
"timestamp": 1709407993000
},
"funder": [
{
"DOI": "10.13039/501100001659",
"doi-asserted-by": "publisher",
"name": "Deutsche Forschungsgemeinschaft"
}
],
"indexed": {
"date-parts": [
[
2024,
3,
3
]
],
"date-time": "2024-03-03T00:13:14Z",
"timestamp": 1709424794482
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
3
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
3,
1
]
],
"date-time": "2024-03-01T00:00:00Z",
"timestamp": 1709251200000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 1,
"start": {
"date-parts": [
[
2024,
3,
2
]
],
"date-time": "2024-03-02T00:00:00Z",
"timestamp": 1709337600000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0166354224000482?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0166354224000482?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "105840",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2024,
3
]
]
},
"published-print": {
"date-parts": [
[
2024,
3
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"article-title": "Verapamil has antiviral activities that target different steps of the influenza virus replication cycle",
"author": "Alam",
"first-page": "121",
"journal-title": "J. Antivir. Antiretrovir.",
"key": "10.1016/j.antiviral.2024.105840_bib1",
"volume": "8",
"year": "2016"
},
{
"DOI": "10.1016/j.antiviral.2011.08.002",
"article-title": "Antiviral activity of the MEK-inhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo",
"author": "Droebner",
"doi-asserted-by": "crossref",
"first-page": "195",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105840_bib2",
"volume": "92",
"year": "2011"
},
{
"DOI": "10.1016/j.coviro.2012.08.004",
"article-title": "The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance",
"author": "Götte",
"doi-asserted-by": "crossref",
"first-page": "644",
"journal-title": "Curr. Opin. Virol.",
"key": "10.1016/j.antiviral.2024.105840_bib3",
"volume": "2",
"year": "2012"
},
{
"DOI": "10.1002/hep.27815",
"article-title": "Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins",
"author": "Hopcraft",
"doi-asserted-by": "crossref",
"first-page": "1059",
"journal-title": "Hepatology",
"key": "10.1016/j.antiviral.2024.105840_bib4",
"volume": "62",
"year": "2015"
},
{
"DOI": "10.1021/acscentsci.3c00538",
"article-title": "Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "1658",
"journal-title": "ACS Cent. Sci.",
"key": "10.1016/j.antiviral.2024.105840_bib5",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.3109/07853890.2011.572905",
"article-title": "Ritonavir-boosted protease inhibitors in HIV therapy",
"author": "Hull",
"doi-asserted-by": "crossref",
"first-page": "375",
"journal-title": "Ann. Med.",
"key": "10.1016/j.antiviral.2024.105840_bib6",
"volume": "43",
"year": "2011"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"article-title": "Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir",
"author": "Iketani",
"doi-asserted-by": "crossref",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105840_bib7",
"volume": "613",
"year": "2023"
},
{
"DOI": "10.1016/j.jcv.2020.104585",
"article-title": "SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies",
"author": "Karamitros",
"doi-asserted-by": "crossref",
"journal-title": "J. Clin. Virol.",
"key": "10.1016/j.antiviral.2024.105840_bib8",
"volume": "131",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2017.06.006",
"article-title": "Emetine inhibits replication of RNA and DNA viruses without generating drug-resistant virus variants",
"author": "Khandelwal",
"doi-asserted-by": "crossref",
"first-page": "196",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105840_bib9",
"volume": "144",
"year": "2017"
},
{
"DOI": "10.1371/journal.pone.0020069",
"article-title": "Potent host-directed small-molecule inhibitors of myxovirus rna-dependent rna-polymerases",
"author": "Krumm",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "PLoS One",
"key": "10.1016/j.antiviral.2024.105840_bib10",
"volume": "6",
"year": "2011"
},
{
"DOI": "10.3389/fmicb.2019.00209",
"article-title": "Inhibitor of sarco/endoplasmic reticulum calcium-ATPase impairs multiple steps of paramyxovirus replication",
"author": "Kumar",
"doi-asserted-by": "crossref",
"journal-title": "Front. Microbiol.",
"key": "10.1016/j.antiviral.2024.105840_bib11",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1128/AAC.00725-11",
"article-title": "Receptor tyrosine kinase inhibitors that block replication of influenza A and other viruses",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "5553",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "10.1016/j.antiviral.2024.105840_bib12",
"volume": "55",
"year": "2011"
},
{
"DOI": "10.1016/j.antiviral.2018.10.022",
"article-title": "MNK1 inhibitor as an antiviral agent suppresses buffalopox virus protein synthesis",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "126",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105840_bib13",
"volume": "160",
"year": "2018"
},
{
"DOI": "10.1371/journal.ppat.1001005",
"article-title": "Quasispecies theory and the behavior of RNA viruses",
"author": "Lauring",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "PLoS Pathog.",
"key": "10.1016/j.antiviral.2024.105840_bib14",
"volume": "6",
"year": "2010"
},
{
"DOI": "10.1016/S0014-5793(04)00108-5",
"article-title": "MEK inhibition impairs influenza B virus propagation without emergence of resistant variants",
"author": "Ludwig",
"doi-asserted-by": "crossref",
"first-page": "37",
"journal-title": "FEBS Lett.",
"key": "10.1016/j.antiviral.2024.105840_bib15",
"volume": "561",
"year": "2004"
},
{
"DOI": "10.1093/cid/ciaa1474",
"article-title": "Emerging RNA-dependent RNA polymerase mutation in a remdesivir-treated B-cell immunodeficient patient with protracted coronavirus disease 2019",
"author": "Martinot",
"doi-asserted-by": "crossref",
"first-page": "E1762",
"journal-title": "Clin. Infect. Dis.",
"key": "10.1016/j.antiviral.2024.105840_bib16",
"volume": "73",
"year": "2021"
},
{
"DOI": "10.1016/j.coviro.2017.10.008",
"article-title": "Genetic bottlenecks in intraspecies virus transmission",
"author": "McCrone",
"doi-asserted-by": "crossref",
"first-page": "20",
"journal-title": "Curr. Opin. Virol.",
"key": "10.1016/j.antiviral.2024.105840_bib17",
"volume": "28",
"year": "2018"
},
{
"DOI": "10.1126/sciadv.ade8778",
"article-title": "Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors",
"author": "Moghadasi",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Adv.",
"key": "10.1016/j.antiviral.2024.105840_bib18",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/sj.onc.1207883",
"article-title": "Lung-specific expression of active Raf kinase results in increased mortality of influenza A virus-infected mice",
"author": "Ölschläger",
"doi-asserted-by": "crossref",
"first-page": "6639",
"journal-title": "Oncogene",
"key": "10.1016/j.antiviral.2024.105840_bib19",
"volume": "23",
"year": "2004"
},
{
"DOI": "10.1038/35060098",
"article-title": "Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade",
"author": "Pleschka",
"doi-asserted-by": "crossref",
"first-page": "301",
"journal-title": "Nat. Cell Biol.",
"key": "10.1016/j.antiviral.2024.105840_bib20",
"volume": "3",
"year": "2001"
},
{
"DOI": "10.1111/cmi.12955",
"article-title": "Late activation of the Raf/MEK/ERK pathway is required for translocation of the respiratory syncytial virus F protein to the plasma membrane and efficient viral replication",
"author": "Preugschas",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Cell Microbiol.",
"key": "10.1016/j.antiviral.2024.105840_bib21",
"volume": "21",
"year": "2019"
},
{
"DOI": "10.1128/AAC.01236-09",
"article-title": "Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors",
"author": "Puyang",
"doi-asserted-by": "crossref",
"first-page": "1981",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "10.1016/j.antiviral.2024.105840_bib22",
"volume": "54",
"year": "2010"
},
{
"article-title": "A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes",
"author": "Sanderson",
"first-page": "1",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105840_bib23",
"year": "2023"
},
{
"article-title": "Multidrug-resistant hepatitis B virus strain in a chronic Turkish patient",
"author": "Sayan",
"first-page": "141",
"journal-title": "Hepat. Mon.",
"key": "10.1016/j.antiviral.2024.105840_bib24",
"volume": "10",
"year": "2010"
},
{
"DOI": "10.3390/pharmaceutics14091776",
"article-title": "The MEK1/2 inhibitor ATR-002 (Zapnometinib) synergistically potentiates the antiviral effect of direct-acting anti-SARS-CoV-2 drugs",
"author": "Schreiber",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Pharmaceutics",
"key": "10.1016/j.antiviral.2024.105840_bib25",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1007/s00018-021-04085-1",
"article-title": "The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses",
"author": "Schreiber",
"doi-asserted-by": "crossref",
"journal-title": "Cell. Mol. Life Sci.",
"key": "10.1016/j.antiviral.2024.105840_bib26",
"volume": "79",
"year": "2022"
},
{
"DOI": "10.1016/j.drudis.2021.05.008",
"article-title": "Drug combination therapy for emerging viral diseases",
"author": "Shyr",
"doi-asserted-by": "crossref",
"first-page": "2367",
"journal-title": "Drug Discov. Today",
"key": "10.1016/j.antiviral.2024.105840_bib27",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.3390/pathogens9090725",
"article-title": "In vitro characterization of multidrug-resistant influenza a(H1n1)pdm09 viruses carrying a dual neuraminidase mutation isolated from immunocompromised patients",
"author": "Takashita",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Pathogens",
"key": "10.1016/j.antiviral.2024.105840_bib28",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1038/cr.2012.129",
"article-title": "Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication",
"author": "Van Der Schaar",
"doi-asserted-by": "crossref",
"first-page": "1576",
"journal-title": "Cell Res.",
"key": "10.1016/j.antiviral.2024.105840_bib29",
"volume": "22",
"year": "2012"
},
{
"DOI": "10.1128/spectrum.03331-22",
"article-title": "Combinations of host- and virus-targeting antiviral drugs confer synergistic suppression of SARS-CoV-2",
"author": "Wagoner",
"doi-asserted-by": "crossref",
"journal-title": "Microbiol. Spectr.",
"key": "10.1016/j.antiviral.2024.105840_bib30",
"volume": "10",
"year": "2022"
}
],
"reference-count": 30,
"references-count": 30,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0166354224000482"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Pharmacology"
],
"subtitle": [],
"title": "The host-targeted antiviral drug Zapnometinib exhibits a high barrier to the development of SARS-CoV-2 resistance",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1016/elsevier_cm_policy"
}