Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection
et al., Virus Evolution, doi:10.1093/ve/veae001, Jan 2024
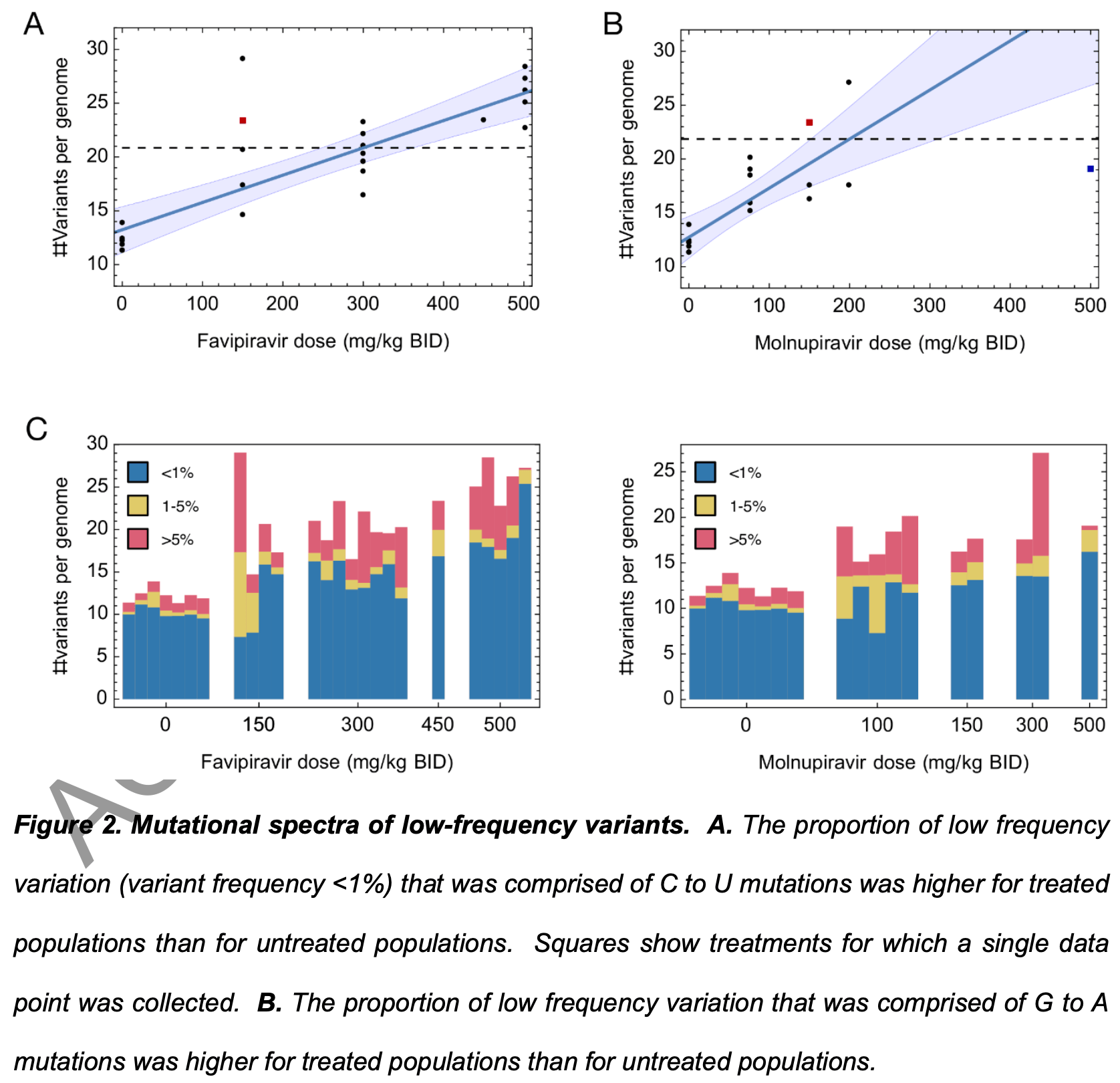
Syrian hamster study showing that short-term treatment with the mutagenic antiviral drugs favipiravir and molnupiravir led to increased genetic variation in SARS-CoV-2 viral populations. Treatment with effective antiviral doses resulted in a gain of 7-10 variants per viral genome compared to untreated controls after only 4 days. The results indicate mutagenic drug treatment can rapidly increase SARS-CoV-2 genetic diversity, which may affect transmission dynamics.
There was a dose-dependent increase in low-frequency mutations that was correlated with reduced viral fitness, suggesting the mutations were deleterious. However, even if the vast majority of mutations caused by these mutagenic drugs are detrimental to the virus, the risk remains that a small fraction could produce dangerous new variants. Just one fitter variant with enhanced pathogenicity or transmissibility could spawn a new wave of infection.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
Study covers molnupiravir and favipiravir.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Illingworth et al., 4 Jan 2024, peer-reviewed, 9 authors.
Contact: christopher.illingworth@glasgow.ac.uk.
Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection
doi:10.1093/ve/veae001/7511244
Mutagenic antiviral drugs have shown promise against multiple viruses, but concerns have been raised about whether their use might promote the emergence of new and harmful viral variants. Recently, genetic signatures associated with molnupiravir use have been identified in the global SARS-COV-2 population. Here, we examine the consequences of using favipiravir and molnupiravir to treat SARS-CoV-2 infection in a hamster model, comparing viral genome sequence data collected from (i) untreated hamsters, and (ii) from hamsters receiving effective
is expected to tend, under mutation-selection balance, to an equilibrium frequency of x=/s, where is the mutation rate and s is the magnitude of selection against the allele 53 , according to the equation Furthermore, assuming mutation rates from one allele to the other are equal, the frequency of a neutral allele is expected to tend to a frequency of one half, according to the equation In both of these formulas, the change in the frequency x is proportional to when x is small. Although positive selection and linkage disequilibrium will affect the evolution of the viral population, we nevertheless sought to fit a linear model to our data.
Analysis of variant composition The proportion of low-frequency variation of distinct mutational classes was measured using variants at frequencies of 1% or below. Briefly, the calculation of q was repeated, considering exclusively low-frequency variants, following which the proportion of this sum that is comprised of each of the 12 mutational classes was calculated. In order to explore the potential adaptive evolution of the viral populations, we identified variants which had reached a frequency of 5% or more. To analyse the composition of these variants we calculated πN/πS, defined as
Non-synonymous and synonymous variation Where cN and cS were the genome-wide counts of non-synonymous and synonymous variants reaching a frequency of 5% or more, and oN and oS are the number of potential nonsynonymous and synonymous variants that..
References
Abdelnabi, Molnupiravir Inhibits Replication of the Emerging SARS-CoV-2 Variants of Concern in a Hamster Infection Model, The Journal of Infectious Diseases
Abdelnabi, The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model, eBioMedicine
Agostini, Small-Molecule Antiviral β-D -N 4 -Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance, J Virol, doi:10.1093/ve/veae001/7511244bygueston06
Alteri, A proof-of-concept study on the genomic evolution of Sars-Cov-2 in molnupiravir-treated, paxlovid-treated and drug-naïve patients, Commun Biol
Bar-On, Flamholz, Phillips, Milo, SARS-CoV-2 (COVID-19) by the numbers, eLife
Bendall, Rapid transmission and tight bottlenecks constrain the evolution of highly transmissible SARS-CoV-2 variants, Nat Commun
Bernal, Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients, N Engl J Med
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, doi:10.1093/ve/veae001/7511244bygueston06
Bull, Sanjuán, Wilke, Theory of Lethal Mutagenesis for Viruses, JVI
Donovan-Banfield, Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial, Nat Commun
Dyer, Covid-19: FDA expert panel recommends authorising molnupiravir but also voices concerns, BMJ
Eloy, Le Grand, Malvy, Guedj, Combined treatment of molnupiravir and favipiravir against SARS-CoV-2 infection: One + zero equals two?, eBioMedicine, doi:10.1093/ve/veae001/7511244bygueston06
Fischer, A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus, Sci. Transl. Med
Gandhi, De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report, Nat Commun
Gao, Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science
Haldane, B S, The Effect of Variation on Fitness, The American Naturalist
Harari, Drivers of adaptive evolution during chronic SARS-CoV-2 infections, Nat Med
Harvey, SARS-CoV-2 variants, spike mutations and immune escape, Nat Rev Microbiol
Hassanipour, The efficacy and safety of Favipiravir in treatment of COVID-19: a systematic review and meta-analysis of clinical trials, Sci Rep
Humphrey, Dalke, Schulten, Vmd, Visual molecular dynamics, Journal of Molecular Graphics
Illingworth, Fitness Inference from Short-Read Data: Within-Host Evolution of a Reassortant H5N1 Influenza Virus, Mol Biol Evol
Illingworth, SAMFIRE: multi-locus variant calling for time-resolved sequence data, Bioinformatics
Jensen, Lynch, Considering mutational meltdown as a potential SARS-CoV-2 treatment strategy, Heredity
Jensen, Stikeleather, Kowalik, Lynch, Imposed mutational meltdown as an antiviral strategy, Evolution
Kabinger, Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat Struct Mol Biol
Kaptein, Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity, Proc. Natl. Acad. Sci
Lauring, Frydman, Andino, The role of mutational robustness in RNA virus evolution, Nat Rev Microbiol, doi:10.1093/ve/veae001/7511244bygueston06
Li, Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics
Li, The Sequence Alignment/Map format and SAMtools, Bioinformatics
Lumby, Favipiravir and Zanamivir Cleared Infection with Influenza B in a Severely Immunocompromised Child, Clinical Infectious Diseases
Lynch, Bürger, Butcher, Gabriel, The Mutational Meltdown in Asexual Populations, Journal of Heredity
Lynch, Conery, Burger, Mutation Accumulation and the Extinction of Small Populations, The American Naturalist
Lythgoe, SARS-CoV-2 within-host diversity and transmission, Science eabg, doi:10.1126/science.abg0821
Mahase, Covid-19: UK becomes first country to authorise antiviral molnupiravir, BMJ
Malone, Campbell, Molnupiravir: coding for catastrophe, Nat Struct Mol Biol
Marshall, A minimal common outcome measure set for COVID-19 clinical research, The Lancet Infectious Diseases
Matuszewski, Ormond, Bank, Jensen, Two sides of the same coin: A population genetics perspective on lethal mutagenesis and mutational meltdown, Virus Evolution
Mccrone, Stochastic processes constrain the within and between host evolution of influenza virus, eLife
Morris, Asynchrony between virus diversity and antibody selection limits influenza virus evolution, eLife, doi:10.1093/ve/veae001/7511244bygueston06
Nelson, Otto, Mutagenic antivirals: the evolutionary risk of low doses
Nguyen, Favipiravir pharmacokinetics in Ebola-Infected patients of the JIKI trial reveals concentrations lower than targeted, PLoS Negl Trop Dis
Painter, The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection, Antiviral Research
Pénisson, Singh, Sniegowski, Gerrish, Dynamics and Fate of Beneficial Mutations Under Lineage Contamination by Linked Deleterious Mutations, Genetics
Reynard, Identification of a New Ribonucleoside Inhibitor of Ebola Virus Replication, Viruses
Ruis, Mutagenesis in Norovirus in Response to Favipiravir Treatment, n engl j med
Sanderson, A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6
Sheahan, An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Sci. Transl. Med
Singh, Singh, Singh, Misra, Molnupiravir in COVID-19: A systematic review of literature, Diabetes & Metabolic Syndrome: Clinical Research & Reviews
Stern, Costs and Benefits of Mutational Robustness in RNA Viruses, Cell Reports
Stevens, Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms, Sci. Transl. Med
Swanstrom, Schinazi, Lethal mutagenesis as an antiviral strategy, Science
Szemiel, In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2, PLoS Pathog
Thorlund, Sheldrick, Meyerowitz-Katz, Singh, Hill, Making Statistical Sense of the Molnupiravir MOVe-OUT Clinical Trial, The American Journal of Tropical Medicine and Hygiene
Wahl, SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801, Nature
Walls, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell
Yoon, Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses, Antimicrob Agents Chemother
DOI record:
{
"DOI": "10.1093/ve/veae001",
"ISSN": [
"2057-1577"
],
"URL": "http://dx.doi.org/10.1093/ve/veae001",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>Mutagenic antiviral drugs have shown promise against multiple viruses, but concerns have been raised about whether their use might promote the emergence of new and harmful viral variants. Recently, genetic signatures associated with molnupiravir use have been identified in the global SARS-COV-2 population. Here, we examine the consequences of using favipiravir and molnupiravir to treat SARS-CoV-2 infection in a hamster model, comparing viral genome sequence data collected from (i) untreated hamsters, and (ii) from hamsters receiving effective and suboptimal doses of treatment. We identify a broadly linear relationship between drug dose and the extent of variation in treated viral populations, with a high proportion of this variation being composed of variants at frequencies of less than one per cent, below typical thresholds for variant calling. Treatment with an effective dose of antiviral drug was associated with a gain of between 7 and 10 variants per viral genome relative to drug-free controls: Even after a short period of treatment a population founded by a transmitted virus could contain multiple sequence differences to that of the original host. Treatment with a suboptimal dose of drug showed intermediate gains of variants. No dose-dependent signal was identified in the numbers of single nucleotide variants reaching frequencies in excess of 5%. We did not find evidence to support the emergence of drug resistance or of novel immune phenotypes. Our study suggests that where onward transmission occurs, a short period of treatment with mutagenic drugs may be sufficient to generate a significant increase in the number of viral variants transmitted.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0002-0030-2784",
"affiliation": [
{
"name": "MRC-University of Glasgow Centre for Virus Research , Glasgow, United Kingdom"
}
],
"authenticated-orcid": false,
"family": "Illingworth",
"given": "Christopher J. R",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Great Ormond Street Hospital for Children NHS Foundation Trust , London, UK"
},
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"family": "Guerra-Assuncao",
"given": "Jose A",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9655-7801",
"affiliation": [
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"authenticated-orcid": false,
"family": "Gregg",
"given": "Samuel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Great Ormond Street Hospital for Children NHS Foundation Trust , London, UK"
},
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"family": "Charles",
"given": "Oscar",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4792-5903",
"affiliation": [
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"authenticated-orcid": false,
"family": "Pang",
"given": "Juanita",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"family": "Roy",
"given": "Sunando",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "KU Leuven Department of Microbiology, Immunology and Transplantation, Rega Institute for Medical Research, Laboratory of Virology and Chemotherapy , B-3000 Leuven, Belgium"
},
{
"name": "The VirusBank Platform, Gaston Geenslaan , B-3000 Leuven, Belgium"
}
],
"family": "Abdelnabi",
"given": "Rana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "KU Leuven Department of Microbiology, Immunology and Transplantation, Rega Institute for Medical Research, Laboratory of Virology and Chemotherapy , B-3000 Leuven, Belgium"
},
{
"name": "The VirusBank Platform, Gaston Geenslaan , B-3000 Leuven, Belgium"
}
],
"family": "Neyts",
"given": "Johan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Great Ormond Street Hospital for Children NHS Foundation Trust , London, UK"
},
{
"name": "Infection, Immunity and Inflammation Research and Teaching Department, University College London , London, UK"
}
],
"family": "Breuer",
"given": "Judith",
"sequence": "additional"
}
],
"container-title": "Virus Evolution",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
1,
5
]
],
"date-time": "2024-01-05T10:27:33Z",
"timestamp": 1704450453000
},
"deposited": {
"date-parts": [
[
2024,
1,
5
]
],
"date-time": "2024-01-05T10:27:34Z",
"timestamp": 1704450454000
},
"funder": [
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_00034/1 MC_UU_12014"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
}
],
"indexed": {
"date-parts": [
[
2024,
1,
6
]
],
"date-time": "2024-01-06T00:17:07Z",
"timestamp": 1704500227474
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
1,
4
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "am",
"delay-in-days": 1,
"start": {
"date-parts": [
[
2024,
1,
5
]
],
"date-time": "2024-01-05T00:00:00Z",
"timestamp": 1704412800000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/ve/advance-article-pdf/doi/10.1093/ve/veae001/55040353/veae001.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/ve/advance-article-pdf/doi/10.1093/ve/veae001/55040353/veae001.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2024,
1,
4
]
]
},
"published-online": {
"date-parts": [
[
2024,
1,
4
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/ve/advance-article/doi/10.1093/ve/veae001/7511244"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Microbiology"
],
"subtitle": [],
"title": "Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection",
"type": "journal-article"
}