The Tautomeric State of N4-Hydroxycytidine within Base-Paired RNA
et al., ACS Central Science, doi:10.1021/acscentsci.4c00146, Apr 2024
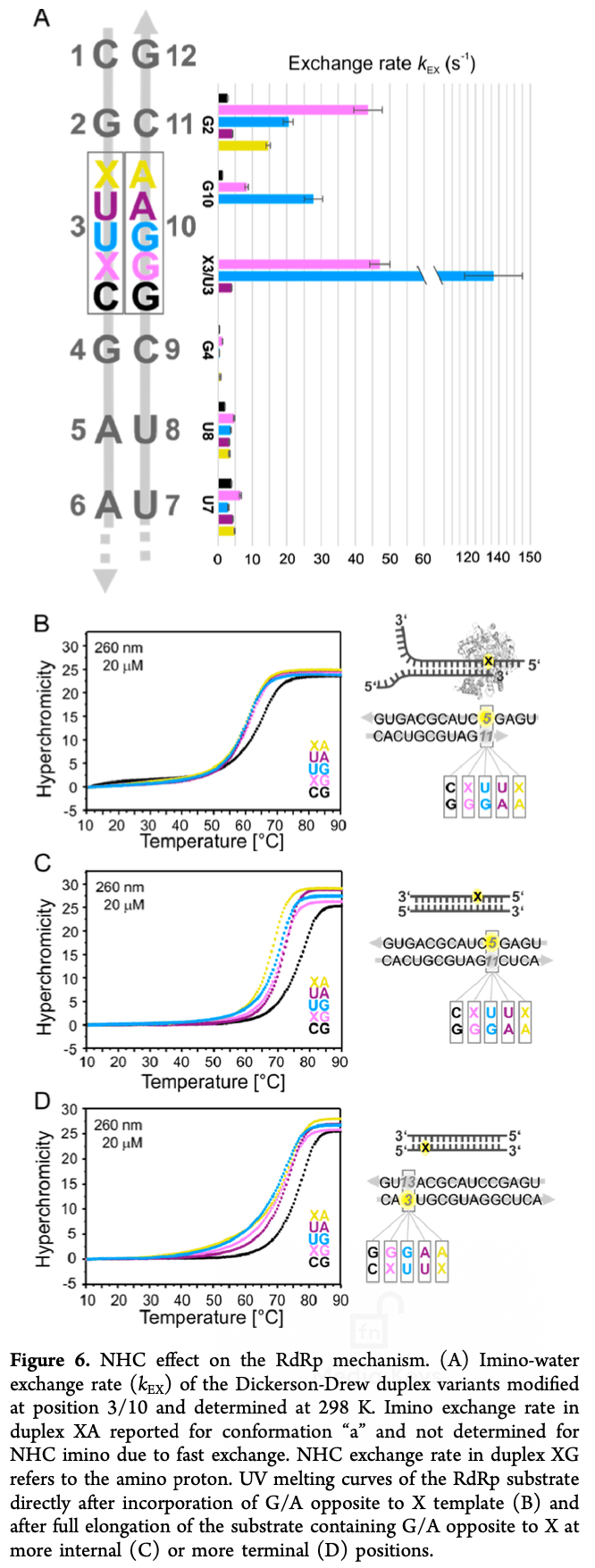
In vitro study showing that NHC, the active form of molnupiravir, can base pair with both G and A in two different tautomeric forms. This ambiguous base pairing enables NHC to induce mutations in the viral RNA. However, it raises the possibility that NHC could also be incorporated into host cell RNA and DNA, potentially causing unwanted mutations in human cells as well. The significant destabilization of RNA duplexes caused by NHC incorporation, especially the impact on neighboring base pairs, suggests it could disrupt the structure and function of not just viral RNA, but any RNA it gets incorporated into, including human cellular RNA involved in normal physiological processes. The fact that NHC:G pairs are more destabilizing than NHC:A pairs and slow down RNA polymerase more also hints that NHC could have complex, sequence-dependent effects that may be hard to predict and control when it comes to off-target incorporation in human cells.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Bessi et al., 25 Apr 2024, peer-reviewed, 7 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
The Tautomeric State of N4-Hydroxycytidine within Base-Paired RNA
ACS Central Science, doi:10.1021/acscentsci.4c00146
Antiviral nucleoside analogues (e.g., Molnupiravir, Remdesivir) played key roles in the treatment of COVID-19 by targeting SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). The nucleoside of Molnupiravir, N 4 -hydroxycytidine (NHC), exists in two tautomeric forms that pair either with G or A within the RdRp active site, causing an accumulation of viral RNA mutations during replication. Detailed insights into the tautomeric states within base pairs and the structural influence of NHC in RNA are still missing. In this study, we investigate the properties of NHC:G and NHC:A base pairs in a self-complementary RNA duplex by UV thermal melting and NMR spectroscopy using atom-specifically 15 N-labeled versions of NHC that were incorporated into oligonucleotides by solid-phase synthesis. NMR analysis revealed that NHC forms a Watson-Crick base pair with G via its amino form, whereas two equally populated conformations were detected for the NHC:A base pair: a weakly hydrogen-bonded Watson-Crick base pair with NHC in the imino form and another conformation with A shifted toward the minor groove. Moreover, we found a variable influence of NHC:G and NHC:A base pairs on the neighboring duplex environment. This study provides conclusive experimental evidence for the existence of two tautomeric forms of NHC within RNA base pairs.
Author Contributions ∥ I.B. and C.S. contributed equally. The manuscript was written through contributions of all authors.
Notes The authors declare no competing financial interest.
References
Agostini, Pruijssers, Chappell, Gribble, Lu et al., Small-molecule antiviral β-D-N 4 -hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance, J. Virol, doi:10.1128/JVI.01348-19
Andersson, Carlsson, Nekoueishahraki, Brath, Erdelyi, Solvent effects on nitrogen chemical shifts, Annu. Rep. NMR Spectrosc, doi:10.1016/bs.arnmr.2015.04.002
Bereiter, Himmelstoss, Renard, Mairhofer, Egger et al., Impact of 3deazapurine nucleobases on RNA properties, Nucleic Acids Res, doi:10.1093/nar/gkab256
Bernal, Gomes Da Silva, Musungaie, Kovalchuk, Gonzalez et al., Study Group, Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients, N. Engl. J. Med, doi:10.1056/NEJMoa2116044
Brown, Hewlins, Schell, The tautomeric state of N 4hydroxy-and of N 4 -amino-cytosine derivatives, J. Chem. Soc. C, doi:10.1039/j39680001925
Butler, Hobbs, Gbinigie, Rahman, Hayward et al., Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial, Lancet, doi:10.1016/S0140-6736(22)02597-1
Cornish, Giedroc, Hennig, Dissecting noncanonical interactions in frameshift-stimulating mRNA pseudoknots, J. Biomol. NMR, doi:10.1007/s10858-006-9033-x
Cornish, Hennig, Giedroc, A loop 2 cytidinestem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated-1 ribosomal frameshifting, Proc. Natl. Acad. Sci. U. S. A, doi:10.1073/pnas.0506166102
Fazakerley, Gdaniec, Sowers, Base-pair induced shifts in the tautomeric equilibrium of a modified DNA base, J. Mol. Biol, doi:10.1006/jmbi.1993.1119
Gdaniec, Ban, Sowers, Fazakerley, Methoxyamine-induced mutagenesis of nucleic acids. A proton NMR study of oligonucleotides containing N 4 -methoxycytosine paired with adenine or guanine, Eur. J. Biochem, doi:10.1111/j.1432-1033.1996.0271r.x
Goddard, Kneller, Manual, None
Gordon, Tchesnokov, Schinazi, Götte, Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template, J. Biol. Chem, doi:10.1016/j.jbc.2021.100770
Hernandez-Santiago, Beltran, Stuyver, Chu, Schinazi, Metabolism of the anti-hepatitis C virus nucleoside β-D-N 4 -hydroxycytidine in different liver cells, Antimicrob. Agents Chemother, doi:10.1128/AAC.48.12.4636-4642.2004
Hwang, Shaka, Water suppression that works -Excitation sculpting using arbitrary wave-forms and pulsed-field gradients, J. Magn. Reson. A, doi:10.1006/jmra.1995.1047
Kabinger, Stiller, Schmitzova, Dienemann, Kokic et al., Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00651-0
Kierdaszuk, Stolarski, Shugar, Hydroxylamine mutagenesis: Observation of inverted Watson Crick base pairing between N 4 -methoxycytosine and adenine with the aid of natural abundance high resolution 15 N NMR Spectroscopy, Eur. J. Biochem, doi:10.1111/j.1432-1033.1983.tb07186.x
Kulinśka, Psoda, Shugar, Mechanism of hydroxylamine mutagenesis: an infrared study of the association in non-polar solutions of 5-methyl-N 4 -hydroxycytosines, Acta Biochim. Pol
Les, Adamowicz, Rode, Structure and conformation of N 4 -hydroxycytosine and N 4 -hydroxy-5-fluorocytosine. A theoretical ab initio study, Biochim. Biophys. Acta, doi:10.1016/0167-4781(93)90240-E
Li, Hilgenfeld, Whitley, De Clercq, Therapeutic strategies for COVID-19: progress and lessons learned, Nat. Rev. Drug Discov, doi:10.1038/s41573-023-00672-y
Lu, Li, Koo, Piccirilli, Efficient synthesis of N 4 -methyl-and N 4 -hydroxycytidine phosphoramidites, Synthesis, doi:10.1055/s-0030-1258170
Markowski, Sullivan, Roberts, Nitrogen-15 nuclear magnetic resonance spectroscopy of some nucleosides and nucleotides, J. Am. Chem. Soc, doi:10.1021/ja00445a009?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Micura, Pils, Höbartner, Grubmayr, Ebert et al., Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion, Nucleic Acids Res, doi:10.1093/nar/29.19.3997
Nedderman, Stone, Williams, Lin, Brown, Molecular basis for methoxyamine-initiated mutagenesis: 1 H nuclear magnetic resonance studies of oligonucleotide duplexes containing base-modified cytosine residues, J. Mol. Biol, doi:10.1006/jmbi.1993.1219
Neuner, Santner, Kreutz, Micura, The ″Speedy″ Synthesis of Atom-Specific 15 N Imino/Amido-Labeled RNA, Chem.-Eur. J, doi:10.1002/chem.201501275
Nixon, Rangan, Kim, Rich, Hoffman et al., Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot, J. Mol. Biol, doi:10.1016/S0022-2836(02)00779-9
Oziminski, Bycul, Thermodynamic and kinetic characteristics of Molnupiravir tautomers and its complexes with RNA purine bases as an explanation of the possible mechanism of action of this novel antiviral medicine: A quantum-chemical study, J. Org. Chem, doi:10.1021/acs.joc.3c01580?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Piotto, Saudek, Sklenar, Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions, J. Biomol. NMR, doi:10.1007/BF02192855
Rangadurai, Kremser, Shi, Kreutz, Al-Hashimi, Direct evidence for (G)O6•••H(2)-N4(C)(+) hydrogen bonding in transient G(syn)-C(+) and G(syn)-m 5 C(+) Hoogsteen base pairs in duplex DNA from cytosine amino nitrogen off-resonance R(1rho) relaxation dispersion measurements, J. Magn. Reson, doi:10.1016/j.jmr.2019.106589
Ruckriegel, Hohmann, Furtig, A Protonated Cytidine Stabilizes the Ligand-Binding Pocket in the PreQ(1) Riboswitch in Thermophilic Bacteria, ChemBioChem, doi:10.1002/cbic.202300228
Sanderson, Hisner, Donovan-Banfield, Hartman, Lochen et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6
Sheahan, Sims, Zhou, Graham, Pruijssers et al., An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Sci. Transl. Med, doi:10.1126/scitranslmed.abb5883
Shugar, Huber, Birnbaum, I Mechanism of hydroxylamine mutagenesis Crystal structure and conformation of 1,5-dimethyl-N 4 -hydroxycytosine, Biochim. Biophys. Acta-Nucleic Acids, doi:10.1016/0005-2787(76)90050-2
Shugar, Kierdaszuk, New light on tautomerism of purines and pyrimidines and its biological and genetic implications, J. Biosciences, doi:10.1007/BF02702764
Sklenar, Bax, Spin-Echo water suppression for the generation of pure-phase two-dimensional NMR spectra, J. Magn. Reson, doi:10.1016/0022-2364(87)90269-1
Spengler, Singer, Effect of tautomeric shift on mutation: N 4 -methoxycytidine forms hydrogen bonds with adenosine in polymers, Biochemistry, doi:10.1021/bi00528a037?urlappend=%3Fref%3DPDF&jav=VoR&rel=cite-as
Stone, Nedderman, Williams, Thoo Lin, Brown, Molecular basis for methoxyamine initiated mutagenesis: 1 H nuclear magnetic resonance studies of base-modified oligodeoxynucleotides, J. Mol. Biol, doi:10.1016/0022-2836(91)90507-3
Strebitzer, Rangadurai, Plangger, Kremser, Juen et al., 5-Oxyacetic acid modification destabilizes double helical stem structures and favors anionic Watson-Crick like cmo 5 U-G Base Pairs, Chem. Eur. J, doi:10.1002/chem.201805077
Ulrich, Akutsu, Doreleijers, Harano, Ioannidis et al., None, Nucleic Acids Res, doi:10.1093/nar/gkm957
Van Meervelt, Moore, Kong Thoo Lin, Brown, Kennard, Molecular and crystal structure of d (CGCGmo4CG): N 4 -methoxycytosine• guanine base-pairs in Z-DNA, J. Mol. Biol, doi:10.1016/0022-2836(90)90398-6
Yoon, Toots, Lee, Lee, Ludeke et al., Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses, Antimicrob. Agents Chemother, doi:10.1128/AAC.00766-18
Zhou, Hill, Sarkar, Tse, Woodburn et al., beta-D-N 4 -hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells, J. Infect. Dis, doi:10.1093/infdis/jiab247
Zibat, Zhang, Dickmanns, Stegmann, Dobbelstein et al., N 4hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786
DOI record:
{
"DOI": "10.1021/acscentsci.4c00146",
"ISSN": [
"2374-7943",
"2374-7951"
],
"URL": "http://dx.doi.org/10.1021/acscentsci.4c00146",
"alternative-id": [
"10.1021/acscentsci.4c00146"
],
"author": [
{
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"family": "Bessi",
"given": "Irene",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"family": "Stiller",
"given": "Carina",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0009-0002-6943-9495",
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"authenticated-orcid": true,
"family": "Schroeder",
"given": "Till",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"family": "Schäd",
"given": "Benedikt",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6543-5385",
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"authenticated-orcid": true,
"family": "Grüne",
"given": "Matthias",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
}
],
"family": "Dietzsch",
"given": "Julia",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4548-2299",
"affiliation": [
{
"name": "Institute of Organic Chemistry, Julius-Maximilians-University Würzburg, Am Hubland, 97074 Würzburg, Bavaria, Germany"
},
{
"name": "Center for Nanosystems Chemistry, Julius-Maximilians-University Würzburg, 97074 Würzburg, Bavaria, Germany"
}
],
"authenticated-orcid": true,
"family": "Höbartner",
"given": "Claudia",
"sequence": "additional"
}
],
"container-title": "ACS Central Science",
"container-title-short": "ACS Cent. Sci.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
4,
25
]
],
"date-time": "2024-04-25T19:52:06Z",
"timestamp": 1714074726000
},
"deposited": {
"date-parts": [
[
2024,
5,
22
]
],
"date-time": "2024-05-22T08:21:59Z",
"timestamp": 1716366119000
},
"funder": [
{
"DOI": "10.13039/501100000781",
"award": [
"682586"
],
"doi-asserted-by": "publisher",
"name": "European Research Council"
},
{
"DOI": "10.13039/501100001659",
"award": [
"277312423",
"46314961"
],
"doi-asserted-by": "publisher",
"name": "Deutsche Forschungsgemeinschaft"
}
],
"indexed": {
"date-parts": [
[
2024,
5,
23
]
],
"date-time": "2024-05-23T00:22:55Z",
"timestamp": 1716423775610
},
"is-referenced-by-count": 0,
"issue": "5",
"issued": {
"date-parts": [
[
2024,
4,
25
]
]
},
"journal-issue": {
"issue": "5",
"published-print": {
"date-parts": [
[
2024,
5,
22
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
4,
25
]
],
"date-time": "2024-04-25T00:00:00Z",
"timestamp": 1714003200000
}
}
],
"link": [
{
"URL": "https://pubs.acs.org/doi/pdf/10.1021/acscentsci.4c00146",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "unspecified"
},
{
"URL": "https://pubs.acs.org/doi/pdf/10.1021/acscentsci.4c00146",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "316",
"original-title": [],
"page": "1084-1093",
"prefix": "10.1021",
"published": {
"date-parts": [
[
2024,
4,
25
]
]
},
"published-online": {
"date-parts": [
[
2024,
4,
25
]
]
},
"published-print": {
"date-parts": [
[
2024,
5,
22
]
]
},
"publisher": "American Chemical Society (ACS)",
"reference": [
{
"DOI": "10.1038/s41573-023-00672-y",
"doi-asserted-by": "publisher",
"key": "ref1/cit1"
},
{
"DOI": "10.1128/AAC.00766-18",
"doi-asserted-by": "publisher",
"key": "ref2/cit2"
},
{
"DOI": "10.1128/JVI.01348-19",
"doi-asserted-by": "publisher",
"key": "ref3/cit3"
},
{
"DOI": "10.1128/AAC.48.12.4636-4642.2004",
"doi-asserted-by": "publisher",
"key": "ref4/cit4"
},
{
"DOI": "10.1056/NEJMoa2116044",
"doi-asserted-by": "publisher",
"key": "ref5/cit5"
},
{
"DOI": "10.1016/S0140-6736(22)02597-1",
"doi-asserted-by": "publisher",
"key": "ref6/cit6"
},
{
"DOI": "10.1093/infdis/jiab247",
"doi-asserted-by": "publisher",
"key": "ref7/cit7"
},
{
"DOI": "10.1016/j.isci.2023.107786",
"doi-asserted-by": "publisher",
"key": "ref8/cit8"
},
{
"DOI": "10.1038/s41586-023-06649-6",
"doi-asserted-by": "publisher",
"key": "ref9/cit9"
},
{
"DOI": "10.1038/s41594-021-00651-0",
"doi-asserted-by": "publisher",
"key": "ref10/cit10"
},
{
"DOI": "10.1016/j.jbc.2021.100770",
"doi-asserted-by": "publisher",
"key": "ref11/cit11"
},
{
"DOI": "10.1126/scitranslmed.abb5883",
"doi-asserted-by": "publisher",
"key": "ref12/cit12"
},
{
"DOI": "10.1039/j39680001925",
"doi-asserted-by": "publisher",
"key": "ref13/cit13"
},
{
"DOI": "10.1007/BF02702764",
"doi-asserted-by": "publisher",
"key": "ref14/cit14"
},
{
"DOI": "10.1016/0005-2787(76)90050-2",
"doi-asserted-by": "publisher",
"key": "ref15/cit15"
},
{
"author": "Kulińska K.",
"first-page": "57",
"issue": "1",
"journal-title": "Acta Biochim. Pol.",
"key": "ref16/cit16",
"volume": "27",
"year": "1980"
},
{
"DOI": "10.1111/j.1432-1033.1983.tb07186.x",
"doi-asserted-by": "publisher",
"key": "ref17/cit17"
},
{
"DOI": "10.1016/0167-4781(93)90240-E",
"doi-asserted-by": "publisher",
"key": "ref18/cit18"
},
{
"DOI": "10.1021/acs.joc.3c01580",
"doi-asserted-by": "publisher",
"key": "ref19/cit19"
},
{
"DOI": "10.1016/0022-2836(90)90398-6",
"doi-asserted-by": "publisher",
"key": "ref20/cit20"
},
{
"DOI": "10.1006/jmbi.1993.1219",
"doi-asserted-by": "publisher",
"key": "ref21/cit21"
},
{
"DOI": "10.1006/jmbi.1993.1119",
"doi-asserted-by": "publisher",
"key": "ref22/cit22"
},
{
"DOI": "10.1016/0022-2836(91)90507-3",
"doi-asserted-by": "publisher",
"key": "ref23/cit23"
},
{
"DOI": "10.1021/bi00528a037",
"doi-asserted-by": "publisher",
"key": "ref24/cit24"
},
{
"DOI": "10.1055/s-0030-1258170",
"doi-asserted-by": "publisher",
"key": "ref25/cit25"
},
{
"DOI": "10.1093/nar/29.19.3997",
"doi-asserted-by": "publisher",
"key": "ref26/cit26"
},
{
"DOI": "10.1002/cbic.202300228",
"doi-asserted-by": "publisher",
"key": "ref27/cit27"
},
{
"DOI": "10.1016/j.jmr.2019.106589",
"doi-asserted-by": "publisher",
"key": "ref28/cit28"
},
{
"DOI": "10.1002/chem.201501275",
"doi-asserted-by": "publisher",
"key": "ref29/cit29"
},
{
"DOI": "10.1021/ja00445a009",
"doi-asserted-by": "publisher",
"key": "ref30/cit30"
},
{
"DOI": "10.1093/nar/gkm957",
"doi-asserted-by": "publisher",
"key": "ref31/cit31"
},
{
"DOI": "10.1016/S0022-2836(02)00779-9",
"doi-asserted-by": "publisher",
"key": "ref32/cit32"
},
{
"DOI": "10.1073/pnas.0506166102",
"doi-asserted-by": "publisher",
"key": "ref33/cit33"
},
{
"DOI": "10.1007/s10858-006-9033-x",
"doi-asserted-by": "publisher",
"key": "ref34/cit34"
},
{
"DOI": "10.1016/bs.arnmr.2015.04.002",
"doi-asserted-by": "publisher",
"key": "ref35/cit35"
},
{
"DOI": "10.1111/j.1432-1033.1996.0271r.x",
"doi-asserted-by": "publisher",
"key": "ref36/cit36"
},
{
"author": "Goddard T.",
"key": "ref37/cit37",
"volume-title": "SPARKY Manual",
"year": "2001"
},
{
"DOI": "10.1016/0022-2364(87)90269-1",
"doi-asserted-by": "publisher",
"key": "ref38/cit38"
},
{
"DOI": "10.1006/jmra.1995.1047",
"doi-asserted-by": "publisher",
"key": "ref39/cit39"
},
{
"DOI": "10.1007/BF02192855",
"doi-asserted-by": "publisher",
"key": "ref40/cit40"
},
{
"DOI": "10.1002/chem.201805077",
"doi-asserted-by": "publisher",
"key": "ref41/cit41"
},
{
"DOI": "10.1093/nar/gkab256",
"doi-asserted-by": "publisher",
"key": "ref42/cit42"
}
],
"reference-count": 42,
"references-count": 42,
"relation": {},
"resource": {
"primary": {
"URL": "https://pubs.acs.org/doi/10.1021/acscentsci.4c00146"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "The Tautomeric State of <i>N</i><sup>4</sup>-Hydroxycytidine within Base-Paired RNA",
"type": "journal-article",
"volume": "10"
}