Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2
et al., Frontiers in Pharmacology, doi:10.3389/fphar.2023.1297924, Dec 2023
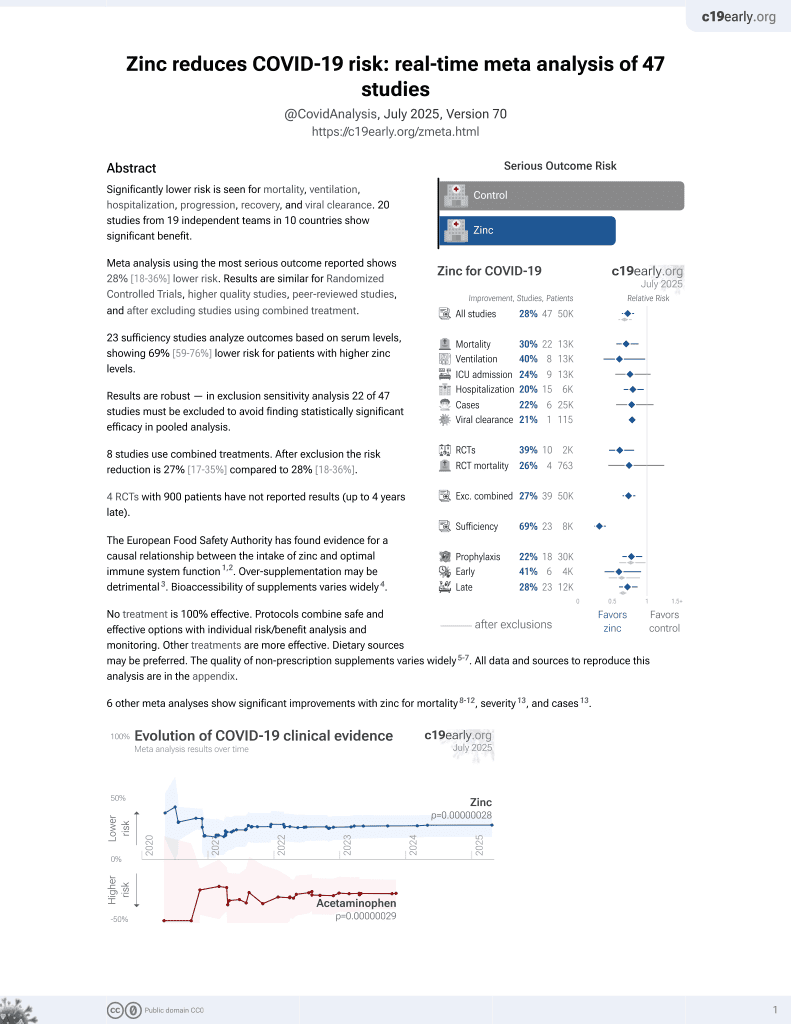
Zinc for COVID-19
2nd treatment shown to reduce risk in
July 2020, now with p = 0.00000028 from 47 studies, recognized in 23 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
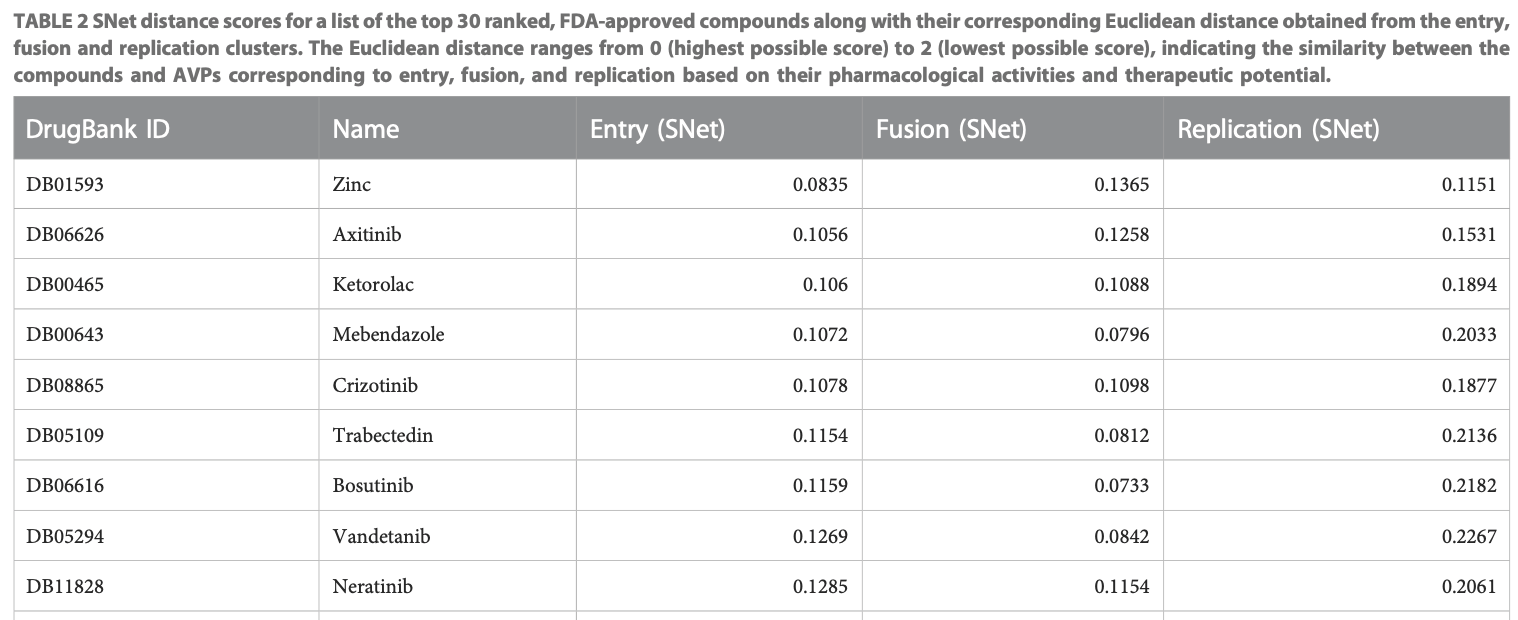
In silico study showing potential benefit of mebendazole and zinc against SARS-CoV-2 according to an AI platform, eVir, designed to identify repurposed oral therapies. The software pipeline analyzes drug impacts on protein-protein networks involved in viral entry, fusion, and replication. Mebendazole and zinc obtained the highest similarity scores to known antiviral peptides.
12 preclinical studies support the efficacy of zinc for COVID-19:
Study covers zinc and mebendazole.
1.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
2.
Lockwood, T., Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent, BioMetals, doi:10.1007/s10534-024-00590-5.
3.
El-Megharbel et al., Chemical and spectroscopic characterization of (Artemisinin/Quercetin/ Zinc) novel mixed ligand complex with assessment of its potent high antiviral activity against SARS-CoV-2 and antioxidant capacity against toxicity induced by acrylamide in male rats, PeerJ, doi:10.7717/peerj.15638.
4.
Bess et al., Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2, Frontiers in Pharmacology, doi:10.3389/fphar.2023.1297924.
5.
Pormohammad et al., Zinc and SARS-CoV-2: A molecular modeling study of Zn interactions with RNA-dependent RNA-polymerase and 3C-like proteinase enzymes, International Journal of Molecular Medicine, doi:10.3892/ijmm.2020.4790.
6.
Pelucelli et al., Zn2+ and Cu2+ Interaction with the Recognition Interface of ACE2 for SARS-CoV-2 Spike Protein, International Journal of Molecular Sciences, doi:10.3390/ijms24119202.
7.
Hajdrik et al., In Vitro Determination of Inhibitory Effects of Humic Substances Complexing Zn and Se on SARS-CoV-2 Virus Replication, Foods, doi:10.3390/foods11050694.
8.
Panchariya et al., Zinc2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro, Chemical Communications, doi:10.1039/D1CC03563K.
Bess et al., 22 Dec 2023, peer-reviewed, 7 authors.
Contact: bess@deepdrug.ai.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2
Frontiers in Pharmacology, doi:10.3389/fphar.2023.1297924
Purpose: This study introduces a sophisticated computational pipeline, eVir, designed for the discovery of antiviral drugs based on their interactions within the human protein network. There is a pressing need for cost-effective therapeutics for infectious diseases (e.g., , particularly in resourcelimited countries. Therefore, our team devised an Artificial Intelligence (AI) system to explore repurposing opportunities for currently used oral therapies. The eVir system operates by identifying pharmaceutical compounds that mirror the effects of antiviral peptides (AVPs)-fragments of human proteins known to interfere with fundamental phases of the viral life cycle: entry, fusion, and replication. eVir extrapolates the probable antiviral efficacy of a given compound by analyzing its established and predicted impacts on the human protein-protein interaction network. This innovative approach provides a promising platform for drug repurposing against SARS-CoV-2 or any virus for which peptide data is available.
Methods: The eVir AI software pipeline processes drug-protein and proteinprotein interaction networks generated from open-source datasets. eVir uses Node2Vec, a graph embedding technique, to understand the nuanced connections among drugs and proteins. The embeddings are input a Siamese Network (SNet) and MLPs, each tailored for the specific mechanisms of entry, fusion, and replication, to evaluate the similarity between drugs and AVPs. Scores generated from the SNet and MLPs undergo a Platt probability calibration and are combined into a unified score that gauges the potential antiviral efficacy of a drug. This integrated approach seeks to boost drug identification confidence, offering a potential solution for detecting therapeutic candidates with pronounced antiviral potency. Once identified a number of compounds were tested for efficacy and toxicity in lung carcinoma cells (Calu-3) infected with SARS-CoV-2. A lead compound was further identified to determine its efficacy and toxicity in K18-hACE2 mice infected with SARS-CoV-2. Computational Predictions: The SNet confidently differentiated between similar and dissimilar drug pairs with an accuracy of 97.28% and AUC of 99.47%.
Ethics statement Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by This animal work was done at IITRI CRO which has appropriate ethics approval. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Conflict of interest Author KW has stock options in Skymount Medical. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abou Dalle, Kantarjian, Burger, Estrov, Ohanian et al., Efficacy and safety of generic imatinib after switching from original imatinib in patients treated for chronic myeloid leukemia in the United States, Cancer Med, doi:10.1002/cam4.2545
Agarwal, Gabrani, Antiviral peptides: identification and validation, Int. J. Peptide Res. Ther, doi:10.1007/s10989-020-10072-0
Amidei, Dobrovolny, Estimation of virus-mediated cell fusion rate of SARS-CoV-2, Virology, doi:10.1016/j.virol.2022.08.016
Ariey-Bonnet, Carrasco, Le Grand, Hoffer, Betzi et al., In silico molecular target prediction unveils mebendazole as a potent MAPK14 inhibitor, Mol. Oncol, doi:10.1002/1878-0261.12810
Assaad, Assaad-Khalil, Imatinib a Tyrosine Kinase Inhibitor: a potential treatment for SARS-COV-2 induced pneumonia, Alexandria J. Med, doi:10.1080/20905068.2020.1778417
Bess, Berglind, Mukhopadhyay, Brylinski, Griggs et al., Artificial intelligence for the discovery of novel antimicrobial agents for emerging infectious diseases, Drug Discov. Today, doi:10.1016/j.drudis.2021.10.022
Bess, None
Bess, None
Bess, None
Bouhaddou, Memon, Meyer, White, Rezelj et al., The global phosphorylation landscape of SARS-CoV-2 infection, Cell, doi:10.1016/j.cell.2020.06.034
Bromley, Bentz, Bottou, Guyon, Lecun et al., Signature verification using a "siamese" time delay neural network, doi:10.1142/s0218001493000339
Dalziel, Round, Garside, Stein, Cost effectiveness of imatinib compared with interferon-alpha or hydroxycarbamide for first-line treatment of chronic myeloid leukaemia, Pharmacoeconomics, doi:10.2165/00019053-200523050-00010
Dittmar, Lee, Whig, Segrist, Li et al., Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2, Cell Rep, doi:10.1016/j.celrep.2021.108959
Drew, Wallingford, Marcotte, MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies, Mol. Syst. Biol, doi:10.15252/msb.202010016
Gancheva, Virchis, Howard-Reeves, Cross, Brazma et al., Myeloproliferative neoplasm with ETV6-ABL1 fusion: a case report and literature review, Mol. Cytogenet, doi:10.1186/1755-8166-6-39
García, Cooper, Shi, Bornmann, Carrion et al., Productive replication of ebola virus is regulated by the c-Abl1 tyrosine kinase, Sci. Transl. Med, doi:10.1126/scitranslmed.3003500
Gargouri, Alzwi, Abobaker, Cyclin dependent kinase inhibitors as a new potential therapeutic option in management of COVID-19, Med. Hypotheses, doi:10.1016/j.mehy.2020.110380
Gassen, Papies, Bajaj, Emanuel, Dethloff et al., SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals, Nat. Commun, doi:10.1038/s41467-021-24007-w
Gilson, Liu, Baitaluk, Nicola, Hwang et al., BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology, Nucleic Acids Res, doi:10.1093/nar/gkv1072
Giurgiu, Reinhard, Brauner, Dunger-Kaltenbach, Fobo et al., CORUM: the comprehensive resource of mammalian protein complexes-2019, Nucleic Acids Res, doi:10.1093/nar/gky973
Grover, Leskovec, Node2vec: scalable feature learning for networks
Hancock, Haney, Gill, The immunology of host defence peptides: beyond antimicrobial activity, Nat. Rev. Immunol, doi:10.1038/nri.2016.29
Kantarjian, Larson, Guilhot, O'brien, Mone et al., Efficacy of imatinib dose escalation in patients with chronic myeloid leukemia in chronic phase, Cancer, doi:10.1002/cncr.24066
Klann, Bojkova, Tascher, Ciesek, Münch et al., Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication, Mol. Cell, doi:10.1016/j.molcel.2020.08.006
Koch, Siamese neural networks for one-shot image recognition
Kuhlmann, Fleckenstein, Antiparasitic agents, Infectious diseases
Lazebnik, Cell fusion as a link between the SARS-CoV-2 spike protein, COVID-19 complications, and vaccine side effects, Oncotarget, doi:10.18632/oncotarget.28088
Liu, Lin, Wen, Jorissen, Gilson, BindingDB: a webaccessible database of experimentally determined protein-ligand binding affinities, Nucleic Acids Res, doi:10.1093/nar/gkl999
Liu, Naderi, Alvin, Mukhopadhyay, Brylinski, Break down in order to build up: decomposing small molecules for fragment-based drug design with eMolFrag, J. Chem. Inf. Model, doi:10.1021/acs.jcim.6b00596
Maxwell, Sanders, Sabot, Hachem, Llanos-Cuentas et al., COVID-19 therapeutics for low-and middle-income countries: a review of candidate agents with potential for near-term use and impact, Am. J. Trop. Med. Hyg, doi:10.4269/ajtmh.21-0200
Mering, Huynen, Jaeggi, Schmidt, Bork et al., STRING: a database of predicted functional associations between proteins, Nucleic Acids Res, doi:10.1093/nar/gkg034
Mikolov, Chen, Corrado, Dean, Efficient estimation of Word representations in vector space
Mikolov, Sutskever, Chen, Corrado, Dean, Distributed representations of words and phrases and their compositionality
Naderi, Alvin, Ding, Mukhopadhyay, Brylinski, A graphbased approach to construct target-focused libraries for virtual screening, J. Cheminform, doi:10.1186/s13321-016-0126-6
Niculescu-Mizil, Caruana, Obtaining calibrated probabilities from boosting
Niculescu-Mizil, Caruana, Predicting good probabilities with supervised learning
Peng, Lloyd, Schran, Clinical pharmacokinetics of imatinib, Clin. Pharmacokinet, doi:10.2165/00003088-200544090-00001
Pickard, Calverley, Chang, Garva, Gago et al., Discovery of re-purposed drugs that slow SARS-CoV-2 replication in human cells, PLoS Pathog, doi:10.1371/journal.ppat.1009840
Pu, Naderi, Liu, Wu, Mukhopadhyay et al., eToxPred: a machine learning-based approach to estimate the toxicity of drug candidates, BMC Pharmacol. Toxicol, doi:10.1186/s40360-018-0282-6
Qureshi, Thakur, Kumar, HIPdb: a database of experimentally validated HIV inhibiting peptides, PLoS One, doi:10.1371/journal.pone.0054908
Qureshi, Thakur, Tandon, Kumar, AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses, Nucleic Acids Res, doi:10.1093/nar/gkt1191
Reeves, Smith, Olson, Thorne, Bornmann et al., Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and Src family tyrosine kinases, J. Virol, doi:10.1128/jvi.01814-10
Sasaki, Ramesh, Chada, Gomyo, Roth et al., The anthelmintic drug mebendazole induces mitotic arrest and apoptosis by depolymerizing tubulin in non-small cell lung cancer cells, Mol. Cancer Ther
Shcherbakov, Ei, Firsova, The mebendazole treatment of patients with alveolar echinococcosis, Med. Parazitol. Mosk
Simbulan-Rosenthal, Dakshanamurthy, Gaur, Chen, Fang et al., The repurposed anthelmintic mebendazole in combination with trametinib suppresses refractory NRASQ61K melanoma, Oncotarget, doi:10.18632/oncotarget.14990
Snel, Lehmann, Bork, Huynen, STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene, Nucleic Acids Res, doi:10.1093/nar/28.18.3442
Szklarczyk, Gable, Lyon, Junge, Wyder et al., STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Res, doi:10.1093/nar/gky1131
Szklarczyk, Kirsch, Koutrouli, Nastou, Mehryary et al., The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest, Nucleic Acids Res, doi:10.1093/nar/gkac1000
Talarico, Pignatti, Pazdur, Registration by the European Agency for the Evaluation of Medicinal products (EMEA) of oncology products approved by the Food and Drug Administration (FDA) under accelerated approval (AA) regulations, J. Clin. Oncol, doi:10.1200/jco.2005.23.16_suppl.6054
Wang, Yang, The therapeutic potential of natural dietary flavonoids against SARS-CoV-2 infection, Nutrients, doi:10.3390/nu15153443
Weston, Coleman, Haupt, Logue, Matthews et al., Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo, J. Virol, doi:10.1128/JVI.01218-20
Wishart, Feunang, Guo, Lo, Marcu et al., DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res, doi:10.1093/nar/gkx1037
Wishart, Knox, Guo, Cheng, Shrivastava et al., DrugBank: a knowledgebase for drugs, drug actions and drug targets, Nucleic Acids Res, doi:10.1093/nar/gkm958
Wishart, Knox, Guo, Shrivastava, Hassanali et al., DrugBank: a comprehensive resource for in silico drug discovery and exploration, Nucleic Acids Res, doi:10.1093/nar/gkj067
Zhao, Mendenhall, Deininger, Imatinib is not a potent anti-SARS-CoV-2 drug, Leukemia, doi:10.1038/s41375-020-01045-9
DOI record:
{
"DOI": "10.3389/fphar.2023.1297924",
"ISSN": [
"1663-9812"
],
"URL": "http://dx.doi.org/10.3389/fphar.2023.1297924",
"abstract": "<jats:p><jats:bold>Purpose:</jats:bold> This study introduces a sophisticated computational pipeline, <jats:italic>eVir</jats:italic>, designed for the discovery of antiviral drugs based on their interactions within the human protein network. There is a pressing need for cost-effective therapeutics for infectious diseases (e.g., COVID-19), particularly in resource-limited countries. Therefore, our team devised an Artificial Intelligence (AI) system to explore repurposing opportunities for currently used oral therapies. The eVir system operates by identifying pharmaceutical compounds that mirror the effects of antiviral peptides (AVPs)—fragments of human proteins known to interfere with fundamental phases of the viral life cycle: entry, fusion, and replication. eVir extrapolates the probable antiviral efficacy of a given compound by analyzing its established and predicted impacts on the human protein-protein interaction network. This innovative approach provides a promising platform for drug repurposing against SARS-CoV-2 or any virus for which peptide data is available.</jats:p><jats:p><jats:bold>Methods:</jats:bold> The <jats:italic>eVir</jats:italic> AI software pipeline processes drug-protein and protein-protein interaction networks generated from open-source datasets. <jats:italic>eVir</jats:italic> uses Node2Vec, a graph embedding technique, to understand the nuanced connections among drugs and proteins. The embeddings are input a Siamese Network (SNet) and MLPs, each tailored for the specific mechanisms of entry, fusion, and replication, to evaluate the similarity between drugs and AVPs. Scores generated from the SNet and MLPs undergo a Platt probability calibration and are combined into a unified score that gauges the potential antiviral efficacy of a drug. This integrated approach seeks to boost drug identification confidence, offering a potential solution for detecting therapeutic candidates with pronounced antiviral potency. Once identified a number of compounds were tested for efficacy and toxicity in lung carcinoma cells (Calu-3) infected with SARS-CoV-2. A lead compound was further identified to determine its efficacy and toxicity in K18-hACE2 mice infected with SARS-CoV-2.</jats:p><jats:p><jats:bold>Computational Predictions:</jats:bold> The SNet confidently differentiated between similar and dissimilar drug pairs with an accuracy of 97.28% and AUC of 99.47%. Key compounds identified through these networks included Zinc, Mebendazole, Levomenol, Gefitinib, Niclosamide, and Imatinib. Notably, Mebendazole and Zinc showcased the highest similarity scores, while Imatinib, Levemenol, and Gefitinib also ranked within the top 20, suggesting their significant pharmacological potentials. Further examination of protein binding analysis using explainable AI focused on reverse engineering the causality of the networks. Protein interaction scores for Mebendazole and Imatinib revealed their effects on notable proteins such as CDPK1, VEGF2, ABL1, and several tyrosine protein kinases.</jats:p><jats:p><jats:bold>Laboratory Studies:</jats:bold> This study determined that Mebendazole, Gefitinib, Topotecan and to some extent Carfilzomib showed conventional drug-response curves, with IC50 values near or below that of Remdesivir with excellent confidence all above R2&gt;0.91, and no cytotoxicity at the IC50 concentration in Calu-3 cells. Cyclosporine A showed antiviral activity, but also unconventional drug-response curves and low R2 which are explained by the non-dose dependent toxicity of the compound. Additionally, Niclosamide demonstrated a conventional drug-response curve with high confidence; however, its inherent cytotoxicity may be a confounding element that misrepresents true antiviral efficacy, by reflecting cellular damage rather than a genuine antiviral action. Remdesivir was used as a control compound and was evaluated in parallel with the submitted test article and had conventional drug-response curves validating the overall results of the assay. Mebendazole was identified from the cell studies to have efficacy at non-toxic concentrations and were further evaluated in mice infected with SARS-CoV-2. Mebendazole administered to K18-hACE2 mice infected with SARS-CoV-2, resulted in a 44.2% reduction in lung viral load compared to non-treated placebo control respectively. There were no significant differences in body weight and all clinical chemistry determinations evaluated (i.e., kidney and liver enzymes) between the different treatment groups.</jats:p><jats:p><jats:bold>Conclusion:</jats:bold> This research underscores the potential of repurposing existing compounds for treating COVID-19. Our preliminary findings underscore the therapeutic promise of several compounds, notably Mebendazole, in both <jats:italic>in vitro</jats:italic> and <jats:italic>in vivo</jats:italic> settings against SARS-CoV-2. Several of the drugs explored, especially Mebendazole, are off-label medication; their cost-effectiveness position them as economical therapies against SARS-CoV-2.</jats:p>",
"alternative-id": [
"10.3389/fphar.2023.1297924"
],
"author": [
{
"affiliation": [],
"family": "Bess",
"given": "Adam",
"sequence": "first"
},
{
"affiliation": [],
"family": "Berglind",
"given": "Frej",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mukhopadhyay",
"given": "Supratik",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Brylinski",
"given": "Michal",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Alvin",
"given": "Chris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fattah",
"given": "Fanan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wasan",
"given": "Kishor M.",
"sequence": "additional"
}
],
"container-title": "Frontiers in Pharmacology",
"container-title-short": "Front. Pharmacol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2023,
12,
22
]
],
"date-time": "2023-12-22T04:42:31Z",
"timestamp": 1703220151000
},
"deposited": {
"date-parts": [
[
2023,
12,
22
]
],
"date-time": "2023-12-22T04:42:36Z",
"timestamp": 1703220156000
},
"indexed": {
"date-parts": [
[
2023,
12,
23
]
],
"date-time": "2023-12-23T00:20:50Z",
"timestamp": 1703290850356
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
12,
22
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
12,
22
]
],
"date-time": "2023-12-22T00:00:00Z",
"timestamp": 1703203200000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fphar.2023.1297924/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2023,
12,
22
]
]
},
"published-online": {
"date-parts": [
[
2023,
12,
22
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.1002/cam4.2545",
"article-title": "Efficacy and safety of generic imatinib after switching from original imatinib in patients treated for chronic myeloid leukemia in the United States",
"author": "Abou Dalle",
"doi-asserted-by": "publisher",
"first-page": "6559",
"journal-title": "Cancer Med.",
"key": "B1",
"volume": "8",
"year": "2019"
},
{
"DOI": "10.1007/s10989-020-10072-0",
"article-title": "Antiviral peptides: identification and validation",
"author": "Agarwal",
"doi-asserted-by": "publisher",
"first-page": "149",
"journal-title": "Int. J. Peptide Res. Ther.",
"key": "B2",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.virol.2022.08.016",
"article-title": "Estimation of virus-mediated cell fusion rate of SARS-CoV-2",
"author": "Amidei",
"doi-asserted-by": "publisher",
"first-page": "91",
"journal-title": "Virology",
"key": "B3",
"volume": "575",
"year": "2022"
},
{
"DOI": "10.1002/1878-0261.12810",
"article-title": "In silico molecular target prediction unveils mebendazole as a potent MAPK14 inhibitor",
"author": "Ariey-Bonnet",
"doi-asserted-by": "publisher",
"first-page": "3083",
"journal-title": "Mol. Oncol.",
"key": "B4",
"volume": "14",
"year": "2020"
},
{
"DOI": "10.1080/20905068.2020.1778417",
"article-title": "Imatinib a Tyrosine Kinase Inhibitor: a potential treatment for SARS- COV-2 induced pneumonia",
"author": "Assaad",
"doi-asserted-by": "publisher",
"first-page": "68",
"journal-title": "Alexandria J. Med.",
"key": "B5",
"volume": "56",
"year": "2020"
},
{
"DOI": "10.1016/j.drudis.2021.10.022",
"article-title": "Artificial intelligence for the discovery of novel antimicrobial agents for emerging infectious diseases",
"author": "Bess",
"doi-asserted-by": "publisher",
"first-page": "1099",
"journal-title": "Drug Discov. Today",
"key": "B6",
"volume": "27",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The global phosphorylation landscape of SARS-CoV-2 infection",
"author": "Bouhaddou",
"doi-asserted-by": "publisher",
"first-page": "685",
"journal-title": "Cell",
"key": "B7",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1142/s0218001493000339",
"article-title": "Signature verification using a “siamese” time delay neural network",
"author": "Bromley",
"doi-asserted-by": "publisher",
"first-page": "669",
"journal-title": "Intern J. Pattern Recognit. Artif. Intell.",
"key": "B8",
"volume": "7",
"year": "1993"
},
{
"key": "B9",
"unstructured": "The safety & efficacy of imatinib for the treatment of SARS-COV-2 induced pneumonia2020"
},
{
"DOI": "10.2165/00019053-200523050-00010",
"article-title": "Cost effectiveness of imatinib compared with interferon-alpha or hydroxycarbamide for first-line treatment of chronic myeloid leukaemia",
"author": "Dalziel",
"doi-asserted-by": "publisher",
"first-page": "515",
"journal-title": "Pharmacoeconomics",
"key": "B10",
"volume": "23",
"year": "2005"
},
{
"DOI": "10.1016/j.celrep.2021.108959",
"article-title": "Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2",
"author": "Dittmar",
"doi-asserted-by": "publisher",
"first-page": "108959",
"journal-title": "Cell Rep.",
"key": "B11",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.15252/msb.202010016",
"article-title": "hu.MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies",
"author": "Drew",
"doi-asserted-by": "publisher",
"first-page": "e10016",
"journal-title": "Mol. Syst. Biol.",
"key": "B12",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1186/1755-8166-6-39",
"article-title": "Myeloproliferative neoplasm with ETV6-ABL1 fusion: a case report and literature review",
"author": "Gancheva",
"doi-asserted-by": "publisher",
"first-page": "39",
"journal-title": "Mol. Cytogenet",
"key": "B13",
"volume": "6",
"year": "2013"
},
{
"DOI": "10.1126/scitranslmed.3003500",
"article-title": "Productive replication of ebola virus is regulated by the c-Abl1 tyrosine kinase",
"author": "García",
"doi-asserted-by": "publisher",
"first-page": "123ra24",
"journal-title": "Sci. Transl. Med.",
"key": "B14",
"volume": "4",
"year": "2012"
},
{
"DOI": "10.1016/j.mehy.2020.110380",
"article-title": "Cyclin dependent kinase inhibitors as a new potential therapeutic option in management of COVID-19",
"author": "Gargouri",
"doi-asserted-by": "publisher",
"first-page": "110380",
"journal-title": "Med. Hypotheses",
"key": "B15",
"volume": "146",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-24007-w",
"article-title": "SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals",
"author": "Gassen",
"doi-asserted-by": "publisher",
"first-page": "3818",
"journal-title": "Nat. Commun.",
"key": "B16",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkv1072",
"article-title": "BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology",
"author": "Gilson",
"doi-asserted-by": "publisher",
"first-page": "D1045",
"journal-title": "Nucleic Acids Res.",
"key": "B17",
"volume": "44",
"year": "2016"
},
{
"DOI": "10.1093/nar/gky973",
"article-title": "CORUM: the comprehensive resource of mammalian protein complexes—2019",
"author": "Giurgiu",
"doi-asserted-by": "publisher",
"first-page": "D559",
"journal-title": "Nucleic Acids Res.",
"key": "B18",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1145/2939672.2939754",
"article-title": "Node2vec: scalable feature learning for networks",
"author": "Grover",
"doi-asserted-by": "crossref",
"key": "B19",
"year": "2016"
},
{
"DOI": "10.1038/nri.2016.29",
"article-title": "The immunology of host defence peptides: beyond antimicrobial activity",
"author": "Hancock",
"doi-asserted-by": "publisher",
"first-page": "321",
"journal-title": "Nat. Rev. Immunol.",
"key": "B20",
"volume": "16",
"year": "2016"
},
{
"DOI": "10.1002/cncr.24066",
"article-title": "Efficacy of imatinib dose escalation in patients with chronic myeloid leukemia in chronic phase",
"author": "Kantarjian",
"doi-asserted-by": "publisher",
"first-page": "551",
"journal-title": "Cancer",
"key": "B21",
"volume": "115",
"year": "2009"
},
{
"DOI": "10.1016/j.molcel.2020.08.006",
"article-title": "Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication",
"author": "Klann",
"doi-asserted-by": "publisher",
"first-page": "164",
"journal-title": "Mol. Cell",
"key": "B22",
"volume": "80",
"year": "2020"
},
{
"article-title": "Siamese neural networks for one-shot image recognition",
"author": "Koch",
"key": "B23",
"year": "2015"
},
{
"DOI": "10.1016/B978-0-7020-6285-8.00157-X",
"article-title": "Antiparasitic agents",
"author": "Kuhlmann",
"doi-asserted-by": "crossref",
"first-page": "1345",
"key": "B24",
"volume-title": "Infectious diseases",
"year": "2017"
},
{
"DOI": "10.18632/oncotarget.28088",
"article-title": "Cell fusion as a link between the SARS-CoV-2 spike protein, COVID-19 complications, and vaccine side effects",
"author": "Lazebnik",
"doi-asserted-by": "publisher",
"first-page": "2476",
"journal-title": "Oncotarget",
"key": "B25",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkl999",
"article-title": "BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities",
"author": "Liu",
"doi-asserted-by": "publisher",
"first-page": "D198",
"journal-title": "Nucleic Acids Res.",
"key": "B26",
"volume": "35",
"year": "2007"
},
{
"DOI": "10.1021/acs.jcim.6b00596",
"article-title": "Break down in order to build up: decomposing small molecules for fragment-based drug design with eMolFrag",
"author": "Liu",
"doi-asserted-by": "publisher",
"first-page": "627",
"journal-title": "J. Chem. Inf. Model",
"key": "B27",
"volume": "57",
"year": "2017"
},
{
"DOI": "10.4269/ajtmh.21-0200",
"article-title": "COVID-19 therapeutics for low- and middle-income countries: a review of candidate agents with potential for near-term use and impact",
"author": "Maxwell",
"doi-asserted-by": "publisher",
"first-page": "584",
"journal-title": "Am. J. Trop. Med. Hyg.",
"key": "B28",
"volume": "105",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkg034",
"article-title": "STRING: a database of predicted functional associations between proteins",
"author": "Mering",
"doi-asserted-by": "publisher",
"first-page": "258",
"journal-title": "Nucleic Acids Res.",
"key": "B29",
"volume": "31",
"year": "2003"
},
{
"article-title": "Efficient estimation of Word representations in vector space",
"author": "Mikolov",
"key": "B30",
"year": ""
},
{
"article-title": "Distributed representations of words and phrases and their compositionality",
"author": "Mikolov",
"key": "B31",
"year": ""
},
{
"DOI": "10.1186/s13321-016-0126-6",
"article-title": "A graph-based approach to construct target-focused libraries for virtual screening",
"author": "Naderi",
"doi-asserted-by": "publisher",
"first-page": "14",
"journal-title": "J. Cheminform",
"key": "B32",
"volume": "8",
"year": "2016"
},
{
"article-title": "Obtaining calibrated probabilities from boosting",
"author": "Niculescu-Mizil",
"key": "B33",
"year": ""
},
{
"DOI": "10.1145/1102351.1102430",
"article-title": "Predicting good probabilities with supervised learning",
"author": "Niculescu-Mizil",
"doi-asserted-by": "crossref",
"key": "B34",
"year": ""
},
{
"DOI": "10.2165/00003088-200544090-00001",
"article-title": "Clinical pharmacokinetics of imatinib",
"author": "Peng",
"doi-asserted-by": "publisher",
"first-page": "879",
"journal-title": "Clin. Pharmacokinet.",
"key": "B35",
"volume": "44",
"year": "2005"
},
{
"DOI": "10.1371/journal.ppat.1009840",
"article-title": "Discovery of re-purposed drugs that slow SARS-CoV-2 replication in human cells",
"author": "Pickard",
"doi-asserted-by": "publisher",
"first-page": "e1009840",
"journal-title": "PLoS Pathog.",
"key": "B36",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1186/s40360-018-0282-6",
"article-title": "eToxPred: a machine learning-based approach to estimate the toxicity of drug candidates",
"author": "Pu",
"doi-asserted-by": "publisher",
"first-page": "2",
"journal-title": "BMC Pharmacol. Toxicol.",
"key": "B37",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1371/journal.pone.0054908",
"article-title": "HIPdb: a database of experimentally validated HIV inhibiting peptides",
"author": "Qureshi",
"doi-asserted-by": "publisher",
"first-page": "e54908",
"journal-title": "PLoS One",
"key": "B38",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.1093/nar/gkt1191",
"article-title": "AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses",
"author": "Qureshi",
"doi-asserted-by": "publisher",
"first-page": "D1147",
"journal-title": "Nucleic Acids Res.",
"key": "B39",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1128/jvi.01814-10",
"article-title": "Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and Src family tyrosine kinases",
"author": "Reeves",
"doi-asserted-by": "publisher",
"first-page": "21",
"journal-title": "J. Virol.",
"key": "B40",
"volume": "85",
"year": "2011"
},
{
"article-title": "The anthelmintic drug mebendazole induces mitotic arrest and apoptosis by depolymerizing tubulin in non-small cell lung cancer cells",
"author": "Sasaki",
"first-page": "1201",
"journal-title": "Mol. Cancer Ther.",
"key": "B41",
"volume": "1",
"year": "2002"
},
{
"article-title": "The mebendazole treatment of patients with alveolar echinococcosis",
"author": "Shcherbakov",
"first-page": "14",
"journal-title": "Med. Parazitol. Mosk.",
"key": "B42",
"year": "1993"
},
{
"DOI": "10.18632/oncotarget.14990",
"article-title": "The repurposed anthelmintic mebendazole in combination with trametinib suppresses refractory NRASQ61K melanoma",
"author": "Simbulan-Rosenthal",
"doi-asserted-by": "publisher",
"first-page": "12576",
"journal-title": "Oncotarget",
"key": "B43",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.1093/nar/28.18.3442",
"article-title": "STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene",
"author": "Snel",
"doi-asserted-by": "publisher",
"first-page": "3442",
"journal-title": "Nucleic Acids Res.",
"key": "B44",
"volume": "28",
"year": "2000"
},
{
"DOI": "10.1093/nar/gky1131",
"article-title": "STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets",
"author": "Szklarczyk",
"doi-asserted-by": "publisher",
"first-page": "D607",
"journal-title": "Nucleic Acids Res.",
"key": "B45",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkac1000",
"article-title": "The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest",
"author": "Szklarczyk",
"doi-asserted-by": "publisher",
"first-page": "D638",
"journal-title": "Nucleic Acids Res.",
"key": "B46",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1200/jco.2005.23.16_suppl.6054",
"article-title": "Registration by the European Agency for the Evaluation of Medicinal products (EMEA) of oncology products approved by the Food and Drug Administration (FDA) under accelerated approval (AA) regulations",
"author": "Talarico",
"doi-asserted-by": "publisher",
"first-page": "6054",
"journal-title": "J. Clin. Oncol.",
"key": "B47",
"volume": "23",
"year": "2005"
},
{
"DOI": "10.3390/nu15153443",
"article-title": "The therapeutic potential of natural dietary flavonoids against SARS-CoV-2 infection",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "3443",
"journal-title": "Nutrients",
"key": "B48",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.1128/JVI.01218-20",
"article-title": "Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo",
"author": "Weston",
"doi-asserted-by": "publisher",
"first-page": "e01218-20",
"journal-title": "J. Virol.",
"key": "B49",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkx1037",
"article-title": "DrugBank 5.0: a major update to the DrugBank database for 2018",
"author": "Wishart",
"doi-asserted-by": "publisher",
"first-page": "D1074",
"journal-title": "Nucleic Acids Res.",
"key": "B50",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.1093/nar/gkm958",
"article-title": "DrugBank: a knowledgebase for drugs, drug actions and drug targets",
"author": "Wishart",
"doi-asserted-by": "publisher",
"first-page": "D901",
"journal-title": "Nucleic Acids Res.",
"key": "B51",
"volume": "36",
"year": "2008"
},
{
"DOI": "10.1093/nar/gkj067",
"article-title": "DrugBank: a comprehensive resource for in silico drug discovery and exploration",
"author": "Wishart",
"doi-asserted-by": "publisher",
"first-page": "D668",
"journal-title": "Nucleic Acids Res.",
"key": "B52",
"volume": "34",
"year": "2006"
},
{
"DOI": "10.1038/s41375-020-01045-9",
"article-title": "Imatinib is not a potent anti-SARS-CoV-2 drug",
"author": "Zhao",
"doi-asserted-by": "publisher",
"first-page": "3085",
"journal-title": "Leukemia",
"key": "B53",
"volume": "34",
"year": "2020"
}
],
"reference-count": 53,
"references-count": 53,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fphar.2023.1297924/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Pharmacology (medical)",
"Pharmacology"
],
"subtitle": [],
"title": "Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.3389/crossmark-policy",
"volume": "14"
}