Genomic Evolution of Sars-Cov-2 in Molnupiravir-Treated Patients Compared to Paxlovid-Treated and Drug-Naïve Patients: A Proof-of-Concept Study
et al., Research Square, doi:10.21203/rs.3.rs-2105569/v1, Oct 2022
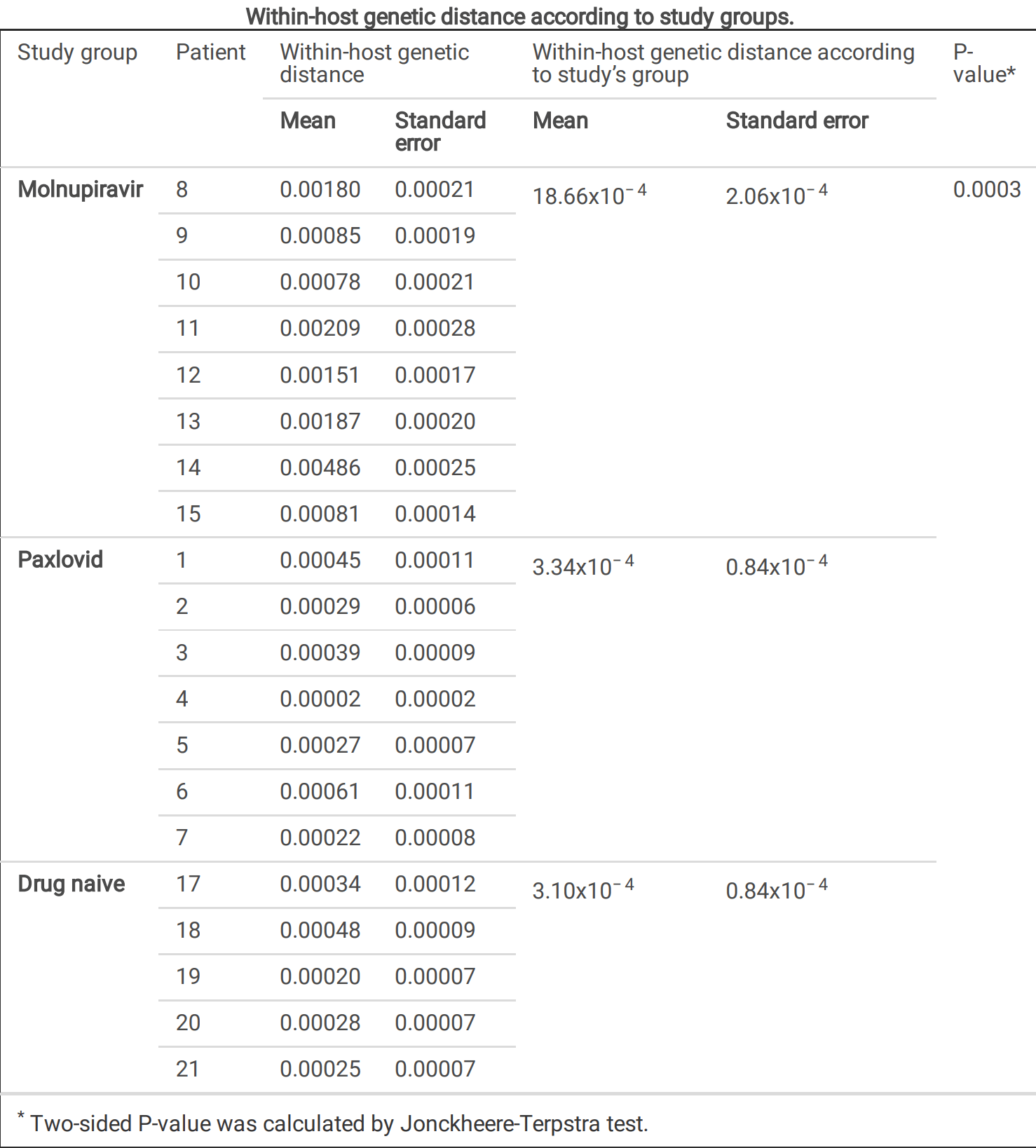
Mutation analysis showing over 5x greater within-host genetic diversity with molnupiravir compared to paxlovid or no treatment.
Potential risks of molnupiravir include the creation of dangerous variants, and mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity1-15. Multiple analyses have identified variants potentially created by molnupiravir16-20. Studies show significantly increased risk of acute kidney injury21, cardiovascular toxocity22, and neurological symptoms21. Treatment may increase viral rebound23,24.
1.
Swanstrom et al., Lethal mutagenesis as an antiviral strategy, Science, doi:10.1126/science.abn0048.
2.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
3.
Shum, C., An investigational study into the drug-associated mutational signature in SARS-CoV-2 viruses, The University of Hong Kong, PhD Thesis, hub.hku.hk/handle/10722/344396.
4.
Waters et al., Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir, Environmental and Molecular Mutagenesis, doi:10.1002/em.22471.
5.
Huntsman, M., An assessment of the reproductive toxicity of the anti-COVID-19 drug molnupiravir using stem cell-based embryo models, Master's Thesis, scholarspace.manoa.hawaii.edu/items/cd11342c-b4dc-44c0-8b44-ce6e3369c40b.
6.
Huntsman (B) et al., Detection of developmental toxicity of the anti-COVID-19 drug molnupiravir using gastruloid-based in vitro assays, Toxicological Sciences, doi:10.1093/toxsci/kfaf093.
7.
Zibat et al., N4-hydroxycytidine, the active compound of Molnupiravir, promotes SARS-CoV-2 mutagenesis and escape from a neutralizing nanobody, iScience, doi:10.1016/j.isci.2023.107786.
8.
Shiraki et al., Convenient screening of the reproductive toxicity of favipiravir and antiviral drugs in Caenorhabditis elegans, Heliyon, doi:10.1016/j.heliyon.2024.e35331.
9.
Gruber et al., Molnupiravir increases SARS‐CoV‐2 genome diversity and complexity: A case‐control cohort study, Journal of Medical Virology, doi:10.1002/jmv.29642.
10.
Marikawa et al., An active metabolite of the anti-COVID-19 drug molnupiravir impairs mouse preimplantation embryos at clinically relevant concentrations, Reproductive Toxicology, doi:10.1016/j.reprotox.2023.108475.
11.
Rahman, M., Elucidation of the DNA repair mechanisms involved in the repair of DNA damage caused by the Arabinosides and Anti-COVID-19 drugs, tokyo-metro-u.repo.nii.ac.jp/records/2000972.
12.
Zhou et al., β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, The Journal of Infectious Diseases, doi:10.1093/infdis/jiab247.
13.
Chamod et al., Molnupiravir Metabolite--N4-hydroxycytidine Causes Cytotoxicity and DNA Damage in Mammalian Cells in vitro: N4-hydroxycytidine Induced Cytotoxicity DNA Damage, Asian Medical Journal and Alternative Medicine, 23:3, asianmedjam.com/index.php/amjam/article/view/1448.
14.
Standing et al., Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients, Nature Communications, doi:10.1038/s41467-024-45641-0.
15.
Mori et al., Reactive oxygen species-mediated cytotoxic and DNA-damaging mechanism of N4-hydroxycytidine, a metabolite of the COVID-19 therapeutic drug molnupiravir, Free Radical Research, doi:10.1080/10715762.2025.2469738.
16.
Focosi et al., The fitness of molnupiravir-signed SARS-CoV-2 variants: imputation analysis based on prescription counts and GISAID analyses by country, Intervirology, doi:10.1159/000540282.
17.
Sanderson et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature, doi:10.1038/s41586-023-06649-6.
18.
Fountain-Jones et al., Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study, The Lancet Microbe, doi:10.1016/S2666-5247(23)00393-2.
19.
Kosakovsky Pond et al., Anti-COVID drug accelerates viral evolution, Nature, doi:10.1038/d41586-023-03248-3.
21.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
22.
Ozhan et al., Evaluation of the cardiopulmonary effects of repurposed COVID-19 therapeutics in healthy rats, Scientific Reports, doi:10.1038/s41598-025-31048-4.
Alteri et al., 6 Oct 2022, retrospective, Italy, preprint, 22 authors, study period March 2022 - May 2022.
Contact: carlofederico.perno@opbg.net.
Genomic Evolution of Sars-Cov-2 in Molnupiravir-Treated Patients Compared to Paxlovid-Treated and Drug-Naïve Patients: A Proof-of-Concept Study
doi:10.21203/rs.3.rs-2105569/v1
Molnupiravir and Paxlovid are the only antivirals approved for COVID-19 treatment. Previous studies have evaluated their e cacy, tolerability, and viral clearance, but little is known about SARS-CoV-2 evolution under their pressure. Here the dynamics of genomic evolution of SARS-CoV-2 in 8 Molnupiravir-treated, 7 Paxlovid-treated and 5 drug-naïve individuals at 4 time-points (Day 0, Day 2, Day 5 of treatment and Day 7) were in-depth investigated. SARS-CoV-2 strains under Molnupiravir pressure were characterized by a higher genetic diversity compared to Paxlovid and no-drug pressure (mean ± SE: 18.66x10 − 4 ±2.06x10 − 4 vs. 3.34x10 − 4 ±0.84x10 − 4 vs. 3.10x10 − 4 ±0.84x10 − 4 , P = 0.0003), with a peak between Day 2 and Day 5. Molnupiravir drove the emergence of more G-A and C-T transitions than other mutations (P = 0.031), regardless of SARS-CoV-2 genes. SARS-CoV-2 under Molnupiravir pressure did not show selective evolution different than that under Paxlovid or no-drug pressure, with the only exception of orf8 (dN > dS, P = 0.001); few amino acid mutations were enriched consistently at speci c sites. No evidence of RdRp or Mpro mutations conferring resistance to Molnupiravir or Paxlovid was found. This proof-of-concept study de nes the SARS-CoV-2 within-host evolution during antiviral treatment, con rming the higher in vivo variability induced by Molnupiravir respect to Paxlovid and controls, albeit not resulting in selection of resistance mutations.
COMPETING INTERESTS The authors have no nancial or non-nancial competing interests that might be perceived to in uence the results and/or discussion reported in this paper.
Supplementary Files This is a list of supplementary les associated with this preprint. Click to download. SupplementaryTable1v.2.0.docx
References
Agostini, Pruijssers, Chappell, Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance, J Virol
Alteri, Cento, Antonello, Detection and quanti cation of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients, PLoS One
Armstrong, Lange, Cesare, Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies, PLoS One
Campos, Pypstra, Rusnak, Investigators, Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19, N Engl J Med
Capriotti, Fariselli, Casadio, I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure, Nucleic Acids Res
Chen, Zhou, Chen, Gu, fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics
El-Kamand, Plessis, Breen, A distinct ssDNA/RNA binding interface in the Nsp9 protein from SARS-CoV-2, Proteins
Fischer Wa 2nd, Eron, Jr, Holman, A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus, Sci Transl Med
Fletcher, Eld, Characterisation of SARS-CoV-2 genomic variations in response to molnupiravir treatment in the AGILE Phase IIa clinical trial, doi:10.21203/rs.3.rs-1835695/v1
Gordon, Tchesnokov, Schinazi, Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template, J Biol Chem
Hillen, Kokic, Farnung, Structure of replicating SARS-CoV-2 polymerase, Nature
Houtgast, Sima, Bertels, Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths, Comput Biol Chem
Imran, Arora, Asdaq, Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19, Molecules
Jungreis, Nelson, Ardern, Con icting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: A homology-based resolution, Virology
Kabinger, Stiller, Schmitzová, Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat Struct Mol Biol
Kalyaanamoorthy, Minh, Wong, ModelFinder: fast model selection for accurate phylogenetic estimates, Nat Methods
Khatib, Benslimane, Elbashir, Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities, Front Cell Infect Microbiol
Kokic, Hillen, Tegunov, Mechanism of SARS-CoV-2 polymerase stalling by remdesivir, Nat Commun
Krzywinski, Schein, Birol, Circos: an information aesthetic for comparative genomics, Genome Res
Lamb, Nirmatrelvir Plus Ritonavir: First Approval, Drugs
Lei, Kusov, Hilgenfeld, Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein, Antiviral Res
Letunic, Bork, Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation, Nucleic Acids Res
Li, A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data, Bioinformatics
Mariano, Farthing, Slm, Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be, Front Mol Biosci
Marzi, Vakil, Bahmanyar, Paxlovid: Mechanism of Action, Synthesis, and In Silico Study, Biomed Res Int
Masyeni, Iqhrammullah, Frediansyah, Molnupiravir: A lethal mutagenic drug against rapidly mutating severe acute respiratory syndrome coronavirus 2-A narrative review, J Med Virol
Minh, Schmidt, Chernomor, IQ-TREE 2: New Models and E cient Methods for Phylogenetic Inference in the Genomic Era, Mol Biol Evol
Moeller, Shi, Demir, Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN
Nei, Gojobori, Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions, Mol Biol Evol
Pienaar, Nelson, A quantitative model of error accumulation during PCR ampli cation, Comput Biol Chem
Pond, Frost, Not so different after all: a comparison of methods for detecting amino acid sites under selection, Mol Biol Evol
Ricciardi, Guarino, Giaquinto, The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle, Nature
Rose, Nolan, Moot, Intra-host site-speci c polymorphisms of SARS-CoV-2 is consistent across multiple samples and methodologies, medRxiv
Shen, Xiao, Kang, Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019, Clin Infect Dis
Shin, Mukherjee, Grewe, Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity, Nature
Singh, Toussi, Hackman, Innovative Randomized Phase I Study and Dosing Regimen Selection to Accelerate and Inform Pivotal COVID-19 Trial of Nirmatrelvir, Clin Pharmacol Ther
Swanstrom, Schinazi, Lethal mutagenesis as an antiviral strategy, Science
Takada, Ueda, Watanabe, Genomic diversity of SARS-CoV-2 can be accelerated by a mutation in the nsp14 gene, bioRxiv
Tamura, Nei, Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees, Mol Biol Evol
Tamura, Stecher, Kumar, MEGA11: Molecular Evolutionary Genetics Analysis Version 11, Mol Biol Evol, doi:10.1093/molbev/msab120
Thakur, Sasi, Pillai, SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines, Front Med
Tian, Pang, Li, Molnupiravir and Its Antiviral Activity Against COVID-19, Front Immunol
Yoon, Toots, Lee, Orally E cacious Broad-Spectrum Ribonucleoside Analog Inhibitor of In uenza and Respiratory Syncytial Viruses, Antimicrob Agents Chemother
Zhang, Chen, Li, The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι, Proc Natl Acad Sci U S A
Zhou, Gammeltoft, Ryberg, Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness in Vitro, bioRxiv
DOI record:
{
"DOI": "10.21203/rs.3.rs-2105569/v1",
"URL": "http://dx.doi.org/10.21203/rs.3.rs-2105569/v1",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>Molnupiravir and Paxlovid are the only antivirals approved for COVID-19 treatment. Previous studies have evaluated their efficacy, tolerability, and viral clearance, but little is known about SARS-CoV-2 evolution under their pressure. Here the dynamics of genomic evolution of SARS-CoV-2 in 8 Molnupiravir-treated, 7 Paxlovid-treated and 5 drug-naïve individuals at 4 time-points (Day 0, Day 2, Day 5 of treatment and Day 7) were in-depth investigated. SARS-CoV-2 strains under Molnupiravir pressure were characterized by a higher genetic diversity compared to Paxlovid and no-drug pressure (mean ± SE: 18.66x10<jats:sup>− 4</jats:sup>±2.06x10<jats:sup>− 4</jats:sup> vs. 3.34x10<jats:sup>− 4</jats:sup>±0.84x10<jats:sup>− 4</jats:sup> vs. 3.10x10<jats:sup>− 4</jats:sup>±0.84x10<jats:sup>− 4</jats:sup>, P = 0.0003), with a peak between Day 2 and Day 5. Molnupiravir drove the emergence of more G-A and C-T transitions than other mutations (P = 0.031), regardless of SARS-CoV-2 genes. SARS-CoV-2 under Molnupiravir pressure did not show selective evolution different than that under Paxlovid or no-drug pressure, with the only exception of orf8 (dN > dS, P = 0.001); few amino acid mutations were enriched consistently at specific sites. No evidence of RdRp or Mpro mutations conferring resistance to Molnupiravir or Paxlovid was found. This proof-of-concept study defines the SARS-CoV-2 within-host evolution during antiviral treatment, confirming the higher in vivo variability induced by Molnupiravir respect to Paxlovid and controls, albeit not resulting in selection of resistance mutations.</jats:p>",
"accepted": {
"date-parts": [
[
2022,
9,
26
]
]
},
"author": [
{
"affiliation": [
{
"name": "Department of Oncology and Hemato-oncology, University of Milan, Milan, Italy and Multimodal Research Area, Bambino Gesù Children Hospital IRCCS, Rome, Italy"
}
],
"family": "Alteri",
"given": "Claudia",
"sequence": "first"
},
{
"affiliation": [],
"family": "Fox",
"given": "Valeria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Scutari",
"given": "Rossana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Burastero",
"given": "Giulia Jole",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Volpi",
"given": "Sara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Faltoni",
"given": "Matteo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fini",
"given": "Vanessa",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Granaglia",
"given": "Annarita",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Esperti",
"given": "Sara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gallerani",
"given": "Altea",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5207-8147",
"affiliation": [],
"authenticated-orcid": false,
"family": "Costabile",
"given": "Valentino",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fontana",
"given": "Beatrice",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Franceschini",
"given": "Erica",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1254-9995",
"affiliation": [
{
"name": "University of Modena and Reggio Emilia"
}
],
"authenticated-orcid": false,
"family": "Meschiari",
"given": "Marianna",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Campana",
"given": "Andrea",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bernardi",
"given": "Stefania",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Villani",
"given": "Alberto",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bernaschi",
"given": "Paola",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Russo",
"given": "Cristina",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5724-3914",
"affiliation": [
{
"name": "University of Modena"
}
],
"authenticated-orcid": false,
"family": "Guaraldi",
"given": "Giovanni",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Modena"
}
],
"family": "Mussini",
"given": "Cristina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Perno",
"given": "Carlo",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
10,
6
]
],
"date-time": "2022-10-06T15:45:55Z",
"timestamp": 1665071155000
},
"deposited": {
"date-parts": [
[
2022,
10,
6
]
],
"date-time": "2022-10-06T15:46:21Z",
"timestamp": 1665071181000
},
"group-title": "In Review",
"indexed": {
"date-parts": [
[
2022,
10,
6
]
],
"date-time": "2022-10-06T16:15:26Z",
"timestamp": 1665072926262
},
"institution": [
{
"name": "Research Square"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2022,
10,
6
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2022,
10,
6
]
],
"date-time": "2022-10-06T00:00:00Z",
"timestamp": 1665014400000
}
}
],
"link": [
{
"URL": "https://www.researchsquare.com/article/rs-2105569/v1",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.researchsquare.com/article/rs-2105569/v1.html",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "8761",
"original-title": [],
"posted": {
"date-parts": [
[
2022,
10,
6
]
]
},
"prefix": "10.21203",
"published": {
"date-parts": [
[
2022,
10,
6
]
]
},
"publisher": "Research Square Platform LLC",
"reference": [
{
"key": "ref1",
"unstructured": "https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain"
},
{
"key": "ref2",
"unstructured": "https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19"
},
{
"DOI": "10.3390/molecules26195795",
"article-title": "Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19",
"author": "Imran M",
"doi-asserted-by": "crossref",
"first-page": "5795",
"issue": "19",
"journal-title": "Molecules",
"key": "ref3",
"unstructured": "Imran, M., Kumar Arora, M., Asdaq, S.M.B., et al.: Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19. Molecules. 24(19), 5795 (2021 Sep)",
"volume": "24"
},
{
"author": "Gordon CJ",
"key": "ref4",
"unstructured": "Gordon, C.J., Tchesnokov, E.P., Schinazi, R.F., et al.: Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template.J Biol Chem. 2021Jul;297(1):100770"
},
{
"article-title": "Molnupiravir and Its Antiviral Activity Against COVID-19",
"author": "Tian L",
"first-page": "855496",
"journal-title": "Front. Immunol.",
"key": "ref5",
"unstructured": "Tian, L., Pang, Z., Li, M., et al.: Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 4, 13:855496 (2022 Apr)",
"volume": "4"
},
{
"author": "Swanstrom R",
"key": "ref6",
"unstructured": "Swanstrom, R., Schinazi, R.F.: Lethal mutagenesis as an antiviral strategy.Science. 2022 Feb4;375(6580):497–498"
},
{
"author": "Kokic G",
"key": "ref7",
"unstructured": "Kokic, G., Hillen, H.S., Tegunov, D., et al.: Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat Commun. 2021 Jan 12;12(1):279"
},
{
"DOI": "10.1038/s41594-021-00651-0",
"article-title": "Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis",
"author": "Kabinger F",
"doi-asserted-by": "crossref",
"first-page": "740",
"issue": "9",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref8",
"unstructured": "Kabinger, F., Stiller, C., Schmitzová, J., et al.: Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 28(9), 740–746 (2021 Sep)",
"volume": "28"
},
{
"author": "Marzi M",
"key": "ref9",
"unstructured": "Marzi, M., Vakil, M.K., Bahmanyar, M., et al.: Paxlovid: Mechanism of Action, Synthesis, and In Silico Study.Biomed Res Int. 2022 Jul7;2022:7341493"
},
{
"DOI": "10.1007/s40265-022-01692-5",
"article-title": "Nirmatrelvir Plus Ritonavir: First Approval",
"author": "Lamb YN",
"doi-asserted-by": "crossref",
"first-page": "585",
"issue": "5",
"journal-title": "Drugs.",
"key": "ref10",
"unstructured": "Lamb, Y.N.: Nirmatrelvir Plus Ritonavir: First Approval. Drugs. 82(5), 585–591 (2022 Apr)",
"volume": "82"
},
{
"DOI": "10.1002/jmv.27730",
"article-title": "Molnupiravir: A lethal mutagenic drug against rapidly mutating severe acute respiratory syndrome coronavirus 2-A narrative review",
"author": "Masyeni S",
"doi-asserted-by": "crossref",
"first-page": "3006",
"issue": "7",
"journal-title": "J. Med. Virol.",
"key": "ref11",
"unstructured": "Masyeni, S., Iqhrammullah, M., Frediansyah, A., et al.: Molnupiravir: A lethal mutagenic drug against rapidly mutating severe acute respiratory syndrome coronavirus 2-A narrative review. J. Med. Virol. 94(7), 3006–3016 (2022 Jul)",
"volume": "94"
},
{
"article-title": "EPIC-HR Investigators. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19",
"author": "Campos A",
"first-page": "1397",
"issue": "15",
"journal-title": "N Engl. J. Med.",
"key": "ref12",
"unstructured": "Campos, A., Pypstra, R., Rusnak, J.M.: EPIC-HR Investigators. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N Engl. J. Med. 14(15), 1397–1408 (2022 Apr)",
"volume": "14"
},
{
"author": "Fischer WA",
"key": "ref13",
"unstructured": "Fischer, W.A. 2, Eron, J.J. Jr., Holman, W., et al.: A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci Transl Med. Jan 19;14(628):eabl7430. (2022)",
"year": "2022"
},
{
"DOI": "10.1002/cpt.2603",
"article-title": "Innovative Randomized Phase I Study and Dosing Regimen Selection to Accelerate and Inform Pivotal COVID-19 Trial of Nirmatrelvir",
"author": "Singh RSP",
"doi-asserted-by": "crossref",
"first-page": "101",
"issue": "1",
"journal-title": "Clin. Pharmacol. Ther.",
"key": "ref14",
"unstructured": "Singh, R.S.P., Toussi, S.S., Hackman, F., et al.: Innovative Randomized Phase I Study and Dosing Regimen Selection to Accelerate and Inform Pivotal COVID-19 Trial of Nirmatrelvir. Clin. Pharmacol. Ther. 112(1), 101–111 (2022 Jul)",
"volume": "112"
},
{
"author": "Yoon JJ",
"key": "ref15",
"unstructured": "Yoon, J.J., Toots, M., Lee, S., et al.: Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses.Antimicrob Agents Chemother. 2018 Jul27;62(8):e00766-18"
},
{
"article-title": "Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance",
"author": "Agostini ML",
"first-page": "e01348",
"issue": "24",
"journal-title": "J. Virol.",
"key": "ref16",
"unstructured": "Agostini, M.L., Pruijssers, A.J., Chappell, J.D., et al.: Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 26(24), e01348–e01319 (2019 Nov)",
"volume": "26"
},
{
"DOI": "10.21203/rs.3.rs-1835695/v1]",
"author": "Fletcher T",
"doi-asserted-by": "publisher",
"key": "ref17",
"unstructured": "Fletcher, T., Donovan-Banfield, I., Penrice-Randal, et al.: Characterisation of SARS-CoV-2 genomic variations in response to molnupiravir treatment in the AGILE Phase IIa clinical trial. 18 July 2022, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-1835695/v1]"
},
{
"author": "Zhou Y",
"key": "ref18",
"unstructured": "Zhou, Y., Gammeltoft, K.A., Ryberg, L.A., et al.: Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness in Vitro. bioRxiv 2022.06.06.494921;"
},
{
"author": "Alteri C",
"key": "ref19",
"unstructured": "Alteri, C., Cento, V., Antonello, M., et al.: Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. Sep 8;15(9):e0236311. (2020)",
"year": "2020"
},
{
"author": "Al Khatib HA",
"key": "ref20",
"unstructured": "Al Khatib, H.A., Benslimane, F.M., Elbashir, I.E., et al.: Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities.Front Cell Infect Microbiol. 2020 Oct6;10:575613"
},
{
"author": "Available",
"key": "ref21",
"unstructured": "Available: at https://www.qiagen.com/us/products/next-generation-sequencing/rna-sequencing/qiaseq-direct-sars-cov-2-kits/"
},
{
"author": "Chen S",
"key": "ref22",
"unstructured": "Chen, S., Zhou, Y., Chen, Y., Gu, J.: fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018 Sep 1;34(17):i884-i890"
},
{
"DOI": "10.1016/j.compbiolchem.2018.03.024",
"article-title": "Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths",
"author": "Houtgast EJ",
"doi-asserted-by": "crossref",
"first-page": "54",
"journal-title": "Comput. Biol. Chem.",
"key": "ref23",
"unstructured": "Houtgast, E.J., Sima, V.M., Bertels, K., et al.: Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths. Comput. Biol. Chem. 75, 54–64 (2018 Aug)",
"volume": "75"
},
{
"DOI": "10.1093/bioinformatics/btr509",
"article-title": "A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data",
"author": "Li H",
"doi-asserted-by": "crossref",
"first-page": "2987",
"journal-title": "Bioinformatics",
"key": "ref24",
"unstructured": "Li, H.: A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27, 2987–2993 (2011)",
"volume": "27",
"year": "2011"
},
{
"DOI": "10.1093/molbev/msab120",
"author": "Tamura K",
"doi-asserted-by": "publisher",
"key": "ref25",
"unstructured": "Tamura, K., Stecher, G., Kumar, S.: MEGA11: Molecular Evolutionary Genetics Analysis Version 11.Mol Biol Evol. 2021 Jun25;38(7):3022–3027. doi: 10.1093/molbev/msab120"
},
{
"article-title": "Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees",
"author": "Tamura K",
"first-page": "512",
"issue": "3",
"journal-title": "Mol. Biol. Evol.",
"key": "ref26",
"unstructured": "Tamura, K., Nei, M.: Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10(3), 512–526 (1993 May)",
"volume": "10"
},
{
"author": "Minh BQ",
"key": "ref27",
"unstructured": "Minh, B.Q., Schmidt, H.A., Chernomor, O., et al.: IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. May 1;37(5):1530–1534. (2020)",
"year": "2020"
},
{
"DOI": "10.1038/nmeth.4285",
"article-title": "ModelFinder: fast model selection for accurate phylogenetic estimates",
"author": "Kalyaanamoorthy S",
"doi-asserted-by": "crossref",
"first-page": "587",
"issue": "6",
"journal-title": "Nat. Methods",
"key": "ref28",
"unstructured": "Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., et al.: ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 14(6), 587–589 (2017 Jun)",
"volume": "14"
},
{
"article-title": "Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation",
"author": "Letunic I",
"first-page": "W293",
"issue": "2",
"journal-title": "Nucleic Acids Res",
"key": "ref29",
"unstructured": "Letunic, I., Bork, P.: Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49(2), W293–W296 (2021 Jul)",
"volume": "49"
},
{
"DOI": "10.1016/j.virol.2021.02.013",
"article-title": "Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: A homology-based resolution",
"author": "Jungreis I",
"doi-asserted-by": "crossref",
"first-page": "145",
"journal-title": "Virology",
"key": "ref30",
"unstructured": "Jungreis, I., Nelson, C.W., Ardern, Z., et al.: Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: A homology-based resolution. Virology. 558, 145–151 (2021 Jun)",
"volume": "558"
},
{
"article-title": "Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions",
"author": "Nei M",
"first-page": "418",
"journal-title": "Mol. Biol. Evol.",
"key": "ref31",
"unstructured": "Nei, M., Gojobori, T.: Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986)",
"volume": "3",
"year": "1986"
},
{
"DOI": "10.1093/molbev/msi105",
"article-title": "Not so different after all: a comparison of methods for detecting amino acid sites under selection",
"author": "Kosakovsky Pond SL",
"doi-asserted-by": "crossref",
"first-page": "1208",
"issue": "5",
"journal-title": "Mol. Biol. Evol.",
"key": "ref32",
"unstructured": "Kosakovsky Pond, S.L., Frost, S.D.: Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22(5), 1208–1222 (2005 May)",
"volume": "22"
},
{
"DOI": "10.1101/gr.092759.109",
"article-title": "Circos: an information aesthetic for comparative genomics",
"author": "Krzywinski M",
"doi-asserted-by": "crossref",
"first-page": "1639",
"issue": "9",
"journal-title": "Genome Res.",
"key": "ref33",
"unstructured": "Krzywinski, M., Schein, J., Birol, I., et al.: Circos: an information aesthetic for comparative genomics. Genome Res. 19(9), 1639–1645 (2009 Sep)",
"volume": "19"
},
{
"author": "Lei J",
"key": "ref34",
"unstructured": "Lei, J., Kusov, Y., Hilgenfeld, R.: Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein.Antiviral Res. 2018Jan;149:58–74"
},
{
"author": "Armstrong LA",
"key": "ref35",
"unstructured": "Armstrong, L.A., Lange, S.M., Dee Cesare, V., et al.: Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies.PLoS One. 2021 Jul16;16(7):e0253364"
},
{
"DOI": "10.1038/s41586-022-04835-6",
"article-title": "The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle",
"author": "Ricciardi S",
"doi-asserted-by": "crossref",
"first-page": "761",
"issue": "7915",
"journal-title": "Nature.",
"key": "ref36",
"unstructured": "Ricciardi, S., Guarino, A.M., Giaquinto, L., et al.: The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 606(7915), 761–768 (2022 Jun)",
"volume": "606"
},
{
"DOI": "10.1002/prot.26205",
"article-title": "A distinct ssDNA/RNA binding interface in the Nsp9 protein from SARS-CoV-2",
"author": "El-Kamand S",
"doi-asserted-by": "crossref",
"first-page": "176",
"issue": "1",
"journal-title": "Proteins.",
"key": "ref37",
"unstructured": "El-Kamand, S., Du Plessis, M.D., Breen, N., et al.: A distinct ssDNA/RNA binding interface in the Nsp9 protein from SARS-CoV-2. Proteins. 90(1), 176–185 (2022 Jan)",
"volume": "90"
},
{
"author": "Zhang Y",
"key": "ref38",
"unstructured": "Zhang, Y., Chen, Y., Li, Y., et al.: The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci U S A. Jun 8;118(23):e2024202118. (2021)",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2368-8",
"article-title": "Structure of replicating SARS-CoV-2 polymerase",
"author": "Hillen HS",
"doi-asserted-by": "crossref",
"first-page": "154",
"issue": "7819",
"journal-title": "Nature.",
"key": "ref39",
"unstructured": "Hillen, H.S., Kokic, G., Farnung, L., et al.: Structure of replicating SARS-CoV-2 polymerase. Nature. 584(7819), 154–156 (2020 Aug)",
"volume": "584"
},
{
"author": "Moeller NH",
"key": "ref40",
"unstructured": "Moeller, N.H., Shi, K., Demir, Ã., et al.: Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. bioRxiv [Preprint]. 2021 Apr 4:2021.04.02.438274."
},
{
"author": "Shen Z",
"key": "ref41",
"unstructured": "Shen, Z., Xiao, Y., Kang, L., et al.: Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019.Clin Infect Dis. 2020 Jul28;71(15):713–720"
},
{
"author": "Rose R",
"key": "ref42",
"unstructured": "Rose, R., Nolan, D.J., Moot, S., et al.: Intra-host site-specific polymorphisms of SARS-CoV-2 is consistent across multiple samples and methodologies. medRxiv 2020.04.24.20078691."
},
{
"author": "Capriotti E",
"key": "ref43",
"unstructured": "Capriotti, E., Fariselli, P., Casadio, R.: I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure.Nucleic Acids Res. (2005). Jul 1;33",
"year": "2005"
},
{
"author": "Shin D",
"key": "ref44",
"unstructured": "Shin, D., Mukherjee, R., Grewe, D., et al.: Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity.Nature. 2020Nov;587(7835):657–662"
},
{
"author": "Mariano G",
"key": "ref45",
"unstructured": "Mariano, G., Farthing, R.J., Lale-Farjat, S.L.M., et al.: Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be.Front Mol Biosci. 2020 Dec17;7:605236"
},
{
"author": "Takada K",
"key": "ref46",
"unstructured": "Takada, K., Ueda, M.T., Watanabe, T., et al.: Genomic diversity of SARS-CoV-2 can be accelerated by a mutation in the nsp14 gene. bioRxiv 2020.12.23.424231."
},
{
"article-title": "SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines",
"author": "Thakur S",
"first-page": "815389",
"journal-title": "Front. Med. (Lausanne)",
"key": "ref47",
"unstructured": "Thakur, S., Sasi, S., Pillai, S.G., et al.: SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines. Front. Med. (Lausanne). 22, 9:815389 (2022 Feb)",
"volume": "22"
},
{
"DOI": "10.1016/j.compbiolchem.2005.11.002",
"article-title": "A quantitative model of error accumulation during PCR amplification",
"author": "Pienaar E",
"doi-asserted-by": "crossref",
"first-page": "102",
"issue": "2",
"journal-title": "Comput. Biol. Chem.",
"key": "ref48",
"unstructured": "Pienaar, E., Theron, M., Nelson, M., et al.: A quantitative model of error accumulation during PCR amplification. Comput. Biol. Chem. 30(2), 102–111 (2006 Apr)",
"volume": "30"
}
],
"reference-count": 48,
"references-count": 48,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.researchsquare.com/article/rs-2105569/v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Genomic Evolution of Sars-Cov-2 in Molnupiravir-Treated Patients Compared to Paxlovid-Treated and Drug-Naïve Patients: A Proof-of-Concept Study",
"type": "posted-content"
}