A genetically based computational drug repurposing framework for rapid identification of candidate compounds: application to COVID-19
et al., medRxiv, doi:10.1101/2025.01.10.25320348, Jan 2025
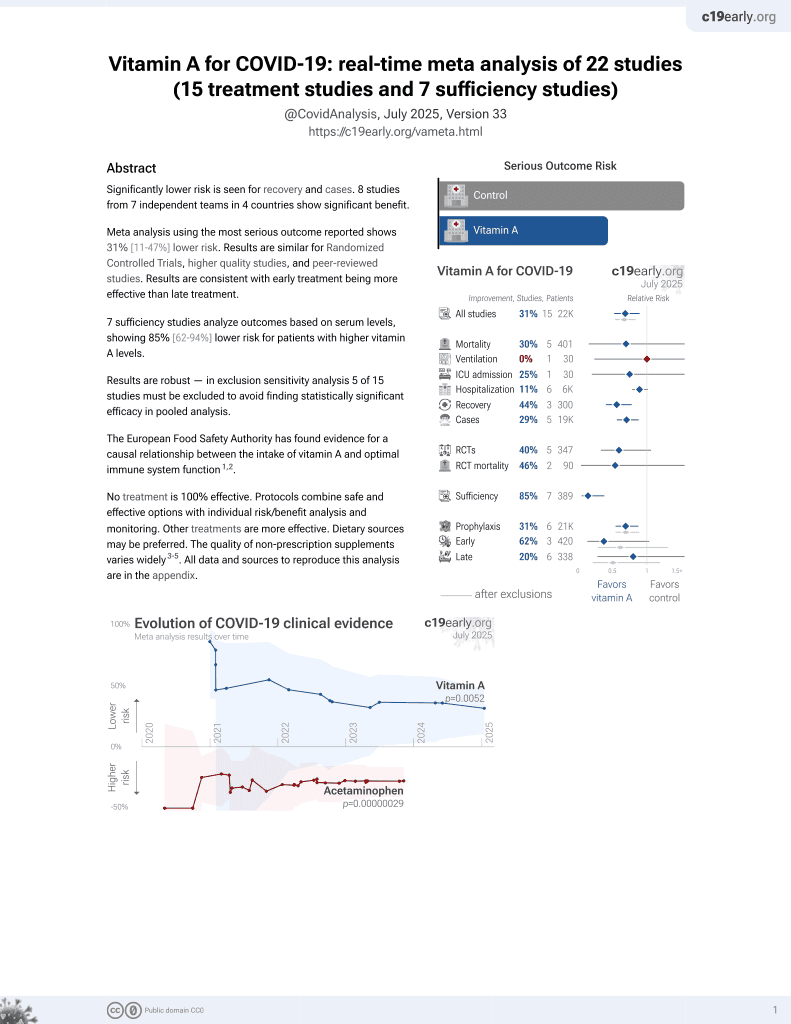
Vitamin A for COVID-19
49th treatment shown to reduce risk in
May 2023, now with p = 0.004 from 14 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Computational drug repurposing study integrating genetically regulated gene expression (GReX) and pharmaceutical databases to identify 7 FDA-approved compounds that may reverse COVID-19-related gene expression.
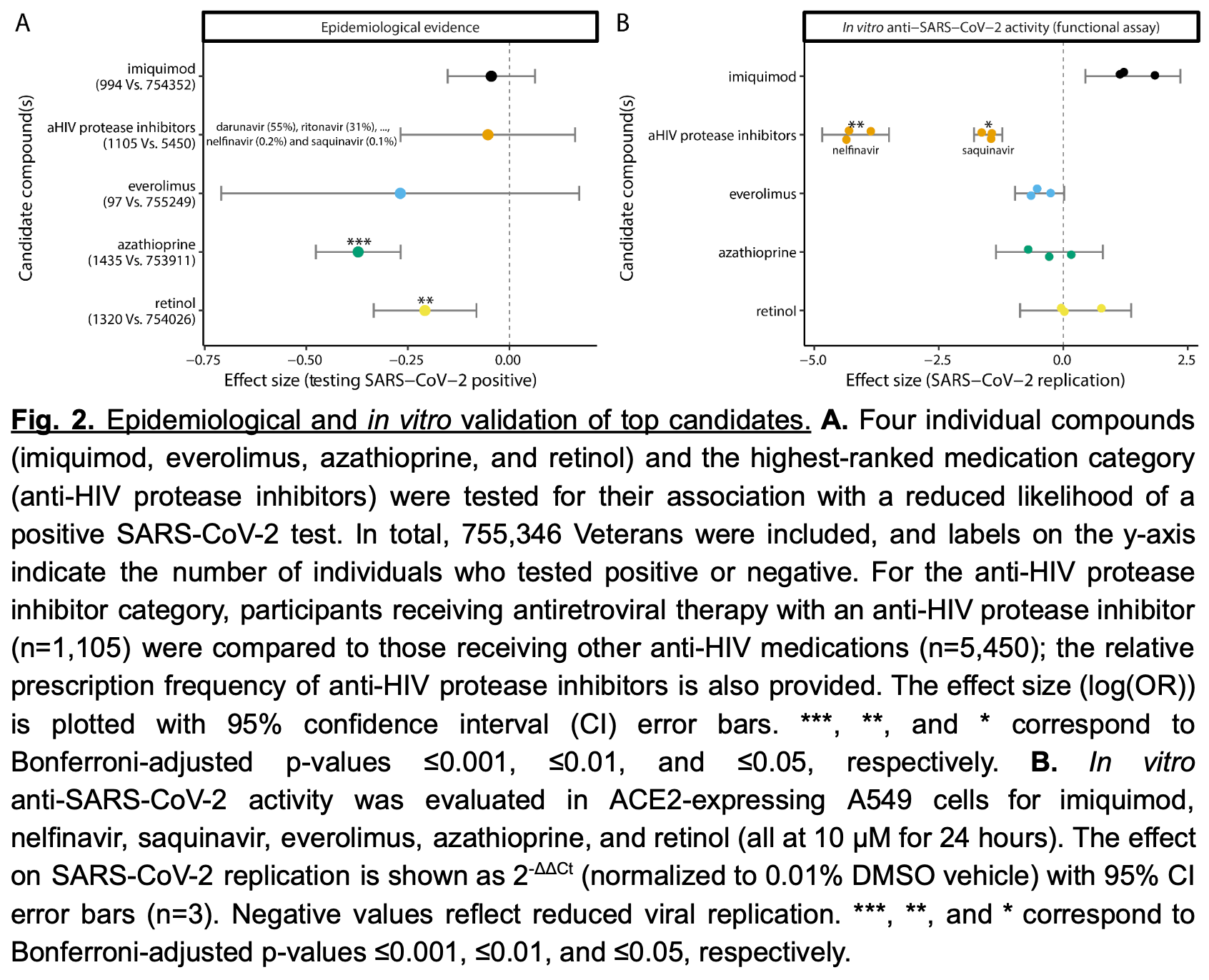
Analysis of 755,346 people in the Veterans Health Administration cohort showed that retinol and azathioprine were associated with reduced incidence of COVID-19.
In vitro analysis showed nelfinavir and saquinavir inhibited SARS-CoV-2 replication by ~95% and ~65% respectively.
11 preclinical studies support the efficacy of vitamin A for COVID-19:
Vitamin A has been identified by the European Food Safety Authority (EFSA) as having sufficient evidence for a causal relationship between intake and optimal immune system function11-13.
Vitamin A has potent antiviral activity against SARS-CoV-2 in both human cell lines and human organoids of the lower respiratory tract (active metabolite all-trans retinoic acid, ATRA)8, is predicted to bind critical host and viral proteins for SARS-CoV-2 and may compensate for gene expression changes related to SARS-CoV-22-4, may be beneficial for COVID-19 via antiviral, anti-inflammatory, and immunomodulatory effects according to network pharmacology analysis5, reduces barrier compromise caused by TNF-α in Calu-3 cells7, inhibits mouse coronavirus replication10, may stimulate innate immunity by activating interferon responses in an IRF3-dependent manner (ATRA)10, may reduce excessive inflammation induced by SARS-CoV-22, shows SARS-CoV-2 antiviral activity In Vitro2,6,9 , is effective against multiple SARS-CoV-2 variants in Calu-3 cells9, and inhibits the entry and replication of SARS-CoV-2 via binding to ACE2 / 3CLpro / RdRp / helicase / 3'-to-5' exonuclease2.
Standard of Care (SOC) for COVID-19 in the study country,
the USA, is very poor with very low average efficacy for approved treatments14.
Only expensive, high-profit treatments were approved for early treatment. Low-cost treatments were excluded, reducing the probability of early treatment due to access and cost barriers, and eliminating complementary and synergistic benefits seen with many low-cost treatments.
|
risk of case, 19.0% lower, OR 0.81, p < 0.001, adjusted per study, multivariable, RR approximated with OR.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
1.
Voloudakis et al., A genetically based computational drug repurposing framework for rapid identification of candidate compounds: application to COVID-19, medRxiv, doi:10.1101/2025.01.10.25320348.
2.
Huang et al., All-trans retinoic acid acts as a dual-purpose inhibitor of SARS-CoV-2 infection and inflammation, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2024.107942.
3.
Chakraborty et al., In-silico screening and in-vitro assay show the antiviral effect of Indomethacin against SARS-CoV-2, Computers in Biology and Medicine, doi:10.1016/j.compbiomed.2022.105788.
4.
Pandya et al., Unravelling Vitamin B12 as a potential inhibitor against SARS-CoV-2: A computational approach, Informatics in Medicine Unlocked, doi:10.1016/j.imu.2022.100951.
5.
Li et al., Revealing the targets and mechanisms of vitamin A in the treatment of COVID-19, Aging, doi:10.18632/aging.103888.
6.
Moatasim et al., Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E, Microorganisms, doi:10.3390/microorganisms11112777.
7.
DiGuilio et al., The multiphasic TNF-α-induced compromise of Calu-3 airway epithelial barrier function, Experimental Lung Research, doi:10.1080/01902148.2023.2193637.
8.
Tong et al., A Retinol Derivative Inhibits SARS-CoV-2 Infection by Interrupting Spike-Mediated Cellular Entry, mBio, doi:10.1128/mbio.01485-22.
9.
Morita et al., All-Trans Retinoic Acid Exhibits Antiviral Effect against SARS-CoV-2 by Inhibiting 3CLpro Activity, Viruses, doi:10.3390/v13081669.
10.
Franco et al., Retinoic Acid-Mediated Inhibition of Mouse Coronavirus Replication Is Dependent on IRF3 and CaMKK, Viruses, doi:10.3390/v16010140.
11.
Galmés et al., Suboptimal Consumption of Relevant Immune System Micronutrients Is Associated with a Worse Impact of COVID-19 in Spanish Populations, Nutrients, doi:10.3390/nu14112254.
12.
Galmés (B) et al., Current State of Evidence: Influence of Nutritional and Nutrigenetic Factors on Immunity in the COVID-19 Pandemic Framework, Nutrients, doi:10.3390/nu12092738.
13.
EFSA, Scientific Opinion on the substantiation of health claims related to vitamin A and cell differentiation (ID 14), function of the immune system (ID 14), maintenance of skin and mucous membranes (ID 15, 17), maintenance of vision (ID 16), maintenance of bone (ID 13, 17), maintenance of teeth (ID 13, 17), maintenance of hair (ID 17), maintenance of nails (ID 17), metabolism of iron (ID 206), and protection of DNA, proteins and lipids from oxidative damage (ID 209) pursuant to Article 13(1) of Regulation (EC) No 1924/2006, EFSA Journal, doi:10.2903/j.efsa.2009.1221.
Voloudakis et al., 14 Jan 2025, retrospective, USA, preprint, 21 authors.
Contact: georgios.voloudakis@mssm.edu, panagiotis.roussos@mssm.edu.
A genetically based computational drug repurposing framework for rapid identification of candidate compounds: application to COVID-19
doi:10.1101/2025.01.10.25320348
Background The development and approval of novel drugs are typically time-intensive and expensive. Leveraging a computational drug repurposing framework that integrates disease-relevant genetically regulated gene expression (GReX) and large longitudinal electronic medical record (EMR) databases can expedite the repositioning of existing medications. However, validating computational predictions of the drug repurposing framework remains a challenge.
Methods To benchmark the drug repurposing framework, we first performed a 5-method-rank-based computational drug prioritization pipeline by integrating multi-tissue GReX associated with COVID-19-related hospitalization, with drug transcriptional signature libraries from the Library of Integrated Network-Based Cellular Signatures. We prioritized FDA-approved medications from the 10 top-ranked compounds, and assessed their association with COVID-19 incidence within the Veterans Health Administration (VHA) cohort (~9 million individuals). In parallel, we evaluated in vitro SARS-CoV-2 replication inhibition in human lung epithelial cells for the selected candidates.
Results Our in silico pipeline identified seven FDA-approved drugs among the top ten candidates. Six (imiquimod, nelfinavir and saquinavir, everolimus, azathioprine, and retinol) had sufficient prescribing rates or feasibility for further testing. In the VHA cohort, azathioprine (odds ratio [OR]=0.69, 95% CI 0.62-0.77) and retinol (OR=0.81, 95% CI 0.72-0.92) were significantly associated with reduced COVID-19 incidence. Conversely, nelfinavir and saquinavir demonstrated potent SARS-CoV-2 inhibition in vitro (~95% and ~65% viral load reduction, respectively). No single compound showed robust protection in both in vivo and in vitro settings. Conclusions These findings underscore the power of GReX-based drug repurposing in rapidly identifying existing therapies with potential clinical relevance; four out of six compounds showed a protective effect in one of the two validation approaches. Crucially, our results highlight how a complementary evaluation-combining epidemiological data and in vitro assays-helps refine
Abbreviations
Abbreviation Definition COVID-19 Coronavirus Disease 2019 SARS-CoV-2 Severe Acute Respiratory Syndrome Coronavirus 2
Supplementary Information Additional File 1: Tables S1-11 Author information Contributions Conceptualization and study design: GV, JFF, JAL, PR. Data contribution or analysis tools: GV, KML, JMV, WZ, DH, SV, JB, MA, ZW, SR, LG, KC, JSL, SKI, S-WL, TLA, GEH, BRtO, JFF, JAL, PR. GV, KML, JMV, WZ, DH, SV, JB performed the analyses. GV, KML, JMV, JFF, JAL, PR wrote the manuscript with input from all authors.
Ethics declarations
Ethics approval and consent to participate Analysis of national VA data was conducted under the protocol, "Leveraging Electronic Health Information to Advance Precision Medicine (LEAP)", which was approved by VA Central Institutional Review Board and by the Research & Development Committees at Palo Alto, Salt Lake City, and West Haven VA Medical Centers.
Consent for publication
Not applicable
Competing interests The authors declare no competing interests.
References
Allegra, Tonacci, Pioggia, Musolino, Gangemi, Vitamin deficiency as risk factor for SARS-CoV-2 infection: with susceptibility and prognosis, Eur Rev Med Pharmacol Sci
Biesheuvel, Vergouwe, Oudega, Hoes, Grobbee et al., Advantages of the nested case-control design in diagnostic research, BMC Med Res Methodol
Blay, Tolani, Ho, Arkin, High-Throughput Screening: today's biochemical and cell-based approaches, Drug Discov Today
Cao, Wang, Wen, Liu, Wang et al., A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19, N Engl J Med
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells, Cell
Desai, Franklin, Alternative approaches for confounding adjustment in observational studies using weighting based on the propensity score: a primer for practitioners, BMJ
Dimasi, Grabowski, Hansen, Innovation in the pharmaceutical industry: New estimates of R&D costs, J Health Econ
Franzén, Ermel, Cohain, Akers, Narzo et al., Cardiometabolic risk loci share downstream cis-and trans-gene regulation across tissues and diseases, Science
Galindez, Matschinske, Rose, Sadegh, Salgado-Albarrán et al., Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies, Nat Comput Sci
Gidari, Sabbatini, Pallotto, Bastianelli, Pierucci et al., Nelfinavir: An Old Ally in the COVID-19 Fight?, Microorganisms
Griffith, Morris, Tudball, Herbert, Mancano et al., Collider bias undermines our understanding of COVID-19 disease risk and severity, Nat Commun
Hochberg, Benjamini, More powerful procedures for multiple significance testing, Stat Med
Hormati, Ghadir, Zamani, Khodadadi, Khodadust et al., Are there any association between COVID-19 severity and immunosuppressive therapy?, Immunol Lett
Li, Wu, Li, Liang, Tse et al., Revealing the targets and mechanisms of vitamin A in the treatment of COVID-19, Aging
Ma, Li, Xie, Zhao, Yi et al., Repurposing of HIV/HCV protease inhibitors against SARS-CoV-2 3CLpro, Antiviral Res
Montazersaheb, Khatibi, Hejazi, Tarhriz, Farjami et al., COVID-19 infection: an overview on cytokine storm and related interventions, Virol J
Nerurkar, Mccoll, Graham, Cavanagh, The systemic response to topical aldara treatment is mediated through direct TLR7 stimulation as imiquimod enters the circulation, Sci Rep
Park, Yu, Kim, Kim, Kim et al., Antiviral Efficacies of FDA-Approved Drugs against SARS-CoV-2 Infection in Ferrets, MBio
Pharmaceuticals, Aldara (imiquimod)
Quan, Sundararajan, Halfon, Fong, Burnand et al., Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data, Med Care
Riva, Yuan, Yin, Martin-Sancho, Matsunaga et al., Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing, Nature
Sarohan, COVID-19: Endogenous Retinoic Acid Theory and Retinoic Acid Depletion Syndrome, Med Hypotheses
Seaman, White, Review of inverse probability weighting for dealing with missing data, Stat Methods Med Res
Shoemark, Colenso, Toelzer, Gupta, Sessions et al., Molecular Simulations suggest Vitamins, Retinoids and Steroids as Ligands of the Free Fatty Acid Pocket of the SARS-CoV-2 Spike Protein*, Angew Chem Int Ed
So, Chau, Chiu, Ho, Lo et al., Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry, Nat Neurosci
Subramanian, Narayan, Corsello, Peck, Natoli et al., A next generation connectivity map: L1000 platform and the first 1,000,000 profiles, Cell
Trasino, A role for retinoids in the treatment of COVID-19?, Clin Exp Pharmacol Physiol
Voloudakis, Vicari, Venkatesh, Hoffman, Dobrindt et al., A translational genomics approach identifies IL10RB as the top candidate gene target for COVID-19 susceptibility, NPJ Genom Med
Xu, Shi, Han, Ling, Jiang et al., Preventive and therapeutic benefits of nelfinavir in rhesus macaques and human beings infected with SARS-CoV-2, Signal Transduct Target Ther
Zhang, Voloudakis, Rajagopal, Readhead, Dudley et al., Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits, Nat Commun
Zong, Moon, Fu, Wang, Zhao, Computational drug repurposing based on electronic health records: a scoping review, npj Digital Med
DOI record:
{
"DOI": "10.1101/2025.01.10.25320348",
"URL": "http://dx.doi.org/10.1101/2025.01.10.25320348",
"abstract": "<jats:p>Background The development and approval of novel drugs are typically time-intensive and expensive. Leveraging a computational drug repurposing framework that integrates disease-relevant genetically regulated gene expression (GReX) and large longitudinal electronic medical record (EMR) databases can expedite the repositioning of existing medications. However, validating computational predictions of the drug repurposing framework remains a challenge. Methods To benchmark the drug repurposing framework, we first performed a 5-method-rank-based computational drug prioritization pipeline by integrating multi-tissue GReX associated with COVID-19-related hospitalization, with drug transcriptional signature libraries from the Library of Integrated Network-Based Cellular Signatures. We prioritized FDA-approved medications from the 10 top-ranked compounds, and assessed their association with COVID-19 incidence within the Veterans Health Administration (VHA) cohort (~9 million individuals). In parallel, we evaluated in vitro SARS-CoV-2 replication inhibition in human lung epithelial cells for the selected candidates. Results Our in silico pipeline identified seven FDA-approved drugs among the top ten candidates. Six (imiquimod, nelfinavir and saquinavir, everolimus, azathioprine, and retinol) had sufficient prescribing rates or feasibility for further testing. In the VHA cohort, azathioprine (odds ratio [OR]=0.69, 95% CI 0.62-0.77) and retinol (OR=0.81, 95% CI 0.72-0.92) were significantly associated with reduced COVID-19 incidence. Conversely, nelfinavir and saquinavir demonstrated potent SARS-CoV-2 inhibition in vitro (~95% and ~65% viral load reduction, respectively). No single compound showed robust protection in both in vivo and in vitro settings. Conclusions These findings underscore the power of GReX-based drug repurposing in rapidly identifying existing therapies with potential clinical relevance; four out of six compounds showed a protective effect in one of the two validation approaches. Crucially, our results highlight how a complementary evaluation-combining epidemiological data and in vitro assays-helps refine the most promising candidates for subsequent mechanistic studies and clinical trials. This integrated validation approach may prove vital for accelerating therapeutic development against current and future health challenges.</jats:p>",
"accepted": {
"date-parts": [
[
2025,
1,
14
]
]
},
"author": [
{
"ORCID": "https://orcid.org/0000-0002-5729-632X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Voloudakis",
"given": "Georgios",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0001-8995-0448",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lee",
"given": "Kyung Min",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4633-9961",
"affiliation": [],
"authenticated-orcid": false,
"family": "Vicari",
"given": "James",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-7370-4306",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhang",
"given": "Wen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6781-3079",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hoagland",
"given": "Daisy",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-3094-2731",
"affiliation": [],
"authenticated-orcid": false,
"family": "Venkatesh",
"given": "Sanan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5989-5177",
"affiliation": [],
"authenticated-orcid": false,
"family": "Bian",
"given": "Jiantao",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0007-8773-0864",
"affiliation": [],
"authenticated-orcid": false,
"family": "Anyfantakis",
"given": "Marios",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0001-4180-7877",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wu",
"given": "Zhenyi",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2744-0965",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rahman",
"given": "Samir",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-9382-0419",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gao",
"given": "Lina",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-1727-7076",
"affiliation": [],
"authenticated-orcid": false,
"family": "Cho",
"given": "Kelly",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lee",
"given": "Jennifer S.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-7488-250X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Iyengar",
"given": "Sudha K.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-8355-5332",
"affiliation": [],
"authenticated-orcid": false,
"family": "Luoh",
"given": "Shiuh-Wen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2349-0009",
"affiliation": [],
"authenticated-orcid": false,
"family": "Assimes",
"given": "Themistocles L.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0957-0224",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hoffman",
"given": "Gabriel",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0324-3078",
"affiliation": [],
"authenticated-orcid": false,
"family": "tenOever",
"given": "Benjamin R.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-9874-2907",
"affiliation": [],
"authenticated-orcid": false,
"family": "Fullard",
"given": "John F.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0108-2127",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lynch",
"given": "Julie A.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4640-6239",
"affiliation": [],
"authenticated-orcid": false,
"family": "Roussos",
"given": "Panos",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
1,
14
]
],
"date-time": "2025-01-14T22:45:19Z",
"timestamp": 1736894719000
},
"deposited": {
"date-parts": [
[
2025,
1,
14
]
],
"date-time": "2025-01-14T22:45:19Z",
"timestamp": 1736894719000
},
"group-title": "Genetic and Genomic Medicine",
"indexed": {
"date-parts": [
[
2025,
1,
15
]
],
"date-time": "2025-01-15T05:32:31Z",
"timestamp": 1736919151720,
"version": "3.33.0"
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
1,
14
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
1,
14
]
],
"date-time": "2025-01-14T00:00:00Z",
"timestamp": 1736812800000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2025.01.10.25320348",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2025,
1,
14
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2025,
1,
14
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2025.01.10.25320348"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "A genetically based computational drug repurposing framework for rapid identification of candidate compounds: application to COVID-19",
"type": "posted-content"
}