Pharmacometrics of high dose ivermectin in early COVID-19: an open label, randomized, controlled adaptive platform trial (PLATCOV)
et al., eLife, doi:10.7554/eLife.83201, PLATCOV, NCT05041907, Jul 2022 (preprint)
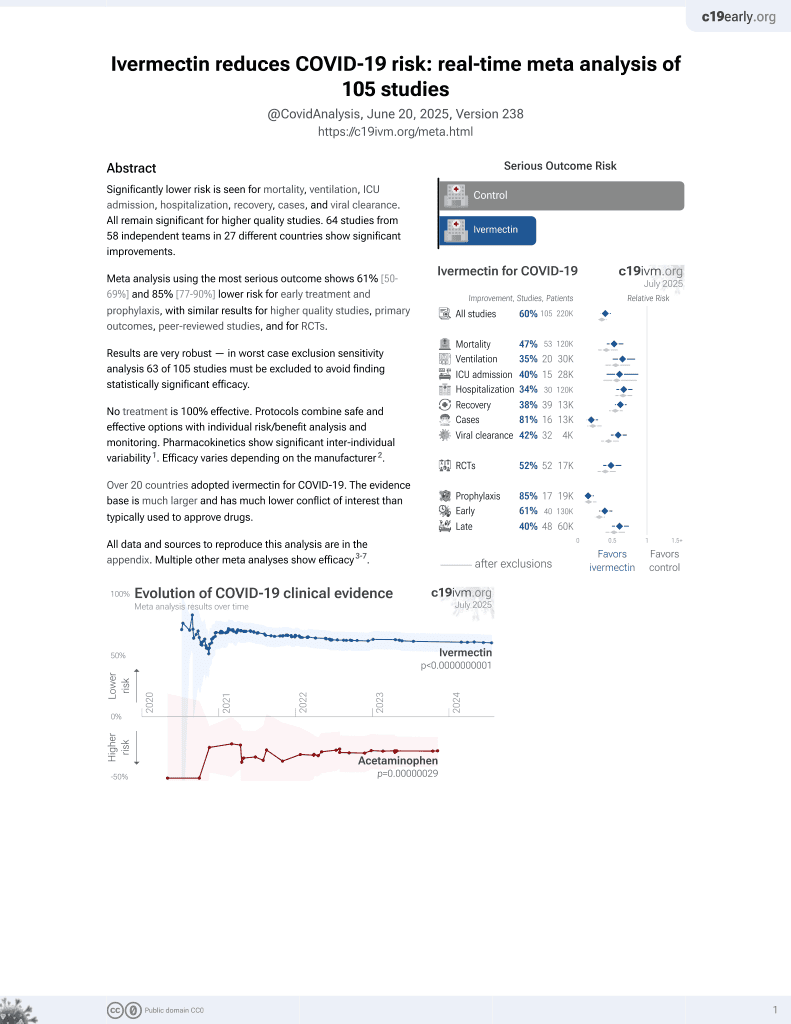
Ivermectin for COVID-19
4th treatment shown to reduce risk in
August 2020, now with p < 0.00000000001 from 106 studies, recognized in 24 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Very high conflict of interest RCT with design optimized for a
null result: very low risk patients, high existing immunity, post-hoc change
to exclude patients more likely to benefit. There was no significant
difference in viral clearance among low risk patients with high viral load at
baseline. All 3 progression events occured in the control arm - one
hospitalization and two cases of COVID-19 related rhabdomyolysis. Patients in
both arms cleared the virus quickly with a viral clearance half-life of 21.1
hours vs. 19.2 hours, which may be in part due to prior immunity.
With rapid viral clearance and very low risk patients,
infection is less likely to spread to other tissues. Systemic treatment is
less applicable, and has less time to reach therapeutic concentrations before
self-recovery.
Treatment administered directly to the respiratory tract, e.g.
as in1-8,
may be more effective for COVID-19 in general, and
extend applicability to fast-resolving cases with infection primarily
localized to the respiratory tract.
The following sentence in the results section, reporting on the
secondary endpoint, was deleted in the journal version: "One patient in the
no study drug arm was hospitalized for clinical reasons before day
28".
Hypothetically, if we want to design a trial to produce a null
effect on viral clearance we can:
•Choose very low risk patients that
typically recover quickly without any treatment.
•Choose patients with high existing
levels of immunity from prior infection or vaccination.
•Make sure we do not test culture
viability and we do not distinguish between live/inactive virus.
While we do not comment on the reason for the design, this
trial does all of these and more with a post-hoc change to exclude patients
from being treated early prior to high viral load:
•Very low risk patients: median age 27,
range 18-45, all patients recovered, no major comorbidities, control arm viral
clearance half-life 0.8 days. This leaves little room for improvement,
especially with an oral treatment. Results with this population have minimal
relevance to real-world usage in patients at higher risk.
•Patients had high existing immunity
with very high vaccination levels and very high baseline antibody positive
results (notably favoring the control arm with 2.2 times as many baseline
antibody negative patients in the ivermectin arm).
•Authors do not test culture viability,
using PCR which does not distinguish between live and disabled virus.
•Authors have made an additional
post-hoc change in favor of finding null results. Specifically, inclusion
criteria were changed from the pre-specified criteria9 to only include
patients with very high viral load. Authors added a restriction to require PCR
Ct <25 or a specific antigen test positive within 2 minutes at baseline. This
minimizes the chance of including patients that may be caught early before
peak viral load, i.e., patients more likely to have benefit from antiviral
mechanisms of action. As shown in Figure S7, almost all patients including
control patients were enrolled at peak viral load and had declining viral load
from baseline. i.e., the selection criteria and population largely prevented
enrolling patients earlier than the point where their immune system was
already efficiently handling the virus.
Note that these methods are synergistic - for example
restricting to high viral load implies greater chance of the virus spreading
to more tissues, requiring longer treatment for oral ivermectin to reach
therapeutic levels in those tissues, thereby reducing the chance of reaching
therapeutic levels before recovery with the population of very fast recovering
patients.
Notably, authors are aware that the post-hoc change favors a
null result — in the discussion they note that results do not apply to
prophylaxis because less potent viral suppression is needed. Similarly, the
required therapeutic level may be much lower as treatment occurs earlier in
the viral cycle when viral load is lower and spread to other tissues is
lower.
Authors provide very little baseline information, however very
large differences are seen - 10, 22, and 50% antibody negative for each arm,
maximum age 31, 45, and 43. Baseline mean viral load was 1.6 times higher in
the ivermectin arm vs. control arm before log conversion.
Figure 2b shows that control patients were enrolled at peak
viral load, while ivermectin patients had peak load one day later. From Figure
S7, this is driven by a small percentage of patients, and may be related to
the 2.2x difference in baseline antibody negative results. These differences
suggest a significant randomization failure in favor of the control group.
Authors provide results for only 10 of the 40
casirivimab/imdevimab patients, which do not match the other arms in terms of
variants, have a much lower maximum age (31 vs. 45), and much lower antibody
negative at baseline (50% versus 22%/10%).
This is the 40th of 53 COVID-19 RCTs for ivermectin, which collectively show efficacy with p=0.000000087.
This is the 89th of 106 COVID-19 controlled studies for ivermectin, which collectively show efficacy with p<0.0000000001.
This study is excluded in the after exclusion results of meta-analysis:
post-hoc change to exclude patients treated before high viral load, population very low risk, recovering quickly without treatment, high baseline immunity, 2.2x greater baseline antibody negative for the treatment arm.
|
risk of hospitalization, 66.7% lower, RR 0.33, p = 1.00, treatment 0 of 45 (0.0%), control 1 of 45 (2.2%), NNT 45, relative risk is not 0 because of continuity correction due to zero events (with reciprocal of the contrasting arm).
|
|
risk of progression, 85.7% lower, RR 0.14, p = 0.24, treatment 0 of 45 (0.0%), control 3 of 45 (6.7%), NNT 15, relative risk is not 0 because of continuity correction due to zero events (with reciprocal of the contrasting arm), hospitalization or progression to COVID-19 rhabdomyolysis.
|
|
relative clearance rate, 9.1% worse, RR 1.09, p = 0.36, treatment 45, control 45, primary outcome.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
1.
Albariqi et al., Pharmacokinetics and Safety of Inhaled Ivermectin in Mice as a Potential COVID-19 Treatment, International Journal of Pharmaceutics, doi:10.1016/j.ijpharm.2022.121688.
2.
Albariqi (B) et al., Preparation and Characterization of Inhalable Ivermectin Powders as a Potential COVID-19 Therapy, Journal of Aerosol Medicine and Pulmonary Drug Delivery, doi:10.1089/jamp.2021.0059.
3.
Chaccour et al., Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, dose-ranging study in rats, Scientific Reports, doi:10.1038/s41598-020-74084-y.
4.
Francés-Monerris et al., Has Ivermectin Virus-Directed Effects against SARS-CoV-2? Rationalizing the Action of a Potential Multitarget Antiviral Agent, ChemRxiv, doi:10.26434/chemrxiv.12782258.v1.
5.
Mansour et al., Safety of inhaled ivermectin as a repurposed direct drug for treatment of COVID-19: A preclinical tolerance study, International Immunopharmacology, doi:10.1016/j.intimp.2021.108004.
6.
Elkholy et al., Ivermectin: A Closer Look at a Potential Remedy, Cureus, doi:10.7759/cureus.10378.
7.
Errecalde et al., Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model, Journal of Pharmaceutical Sciences, doi:10.1016/j.xphs.2021.01.017.
Schilling et al., 19 Jul 2022, Randomized Controlled Trial, Thailand, peer-reviewed, median age 27.0, 38 authors, study period 30 September, 2021 - 18 April, 2022, average treatment delay 2.0 days, dosage 600μg/kg days 1-7, trial NCT05041907 (history) (PLATCOV).
Contact: william@tropmedres.ac, nickw@tropmedres.ac.
Pharmacometrics of high dose ivermectin in early COVID-19: an open label, randomized, controlled adaptive platform trial (PLATCOV)
High dose ivermectin did not have measurable antiviral activity in early symptomatic COVID-19. Pharmacometric evaluation of viral clearance rates based on frequent oropharyngeal sampling is a highly efficient and well-tolerated method of assessing and comparing SARS CoV-2 antiviral therapeutics in vivo.
Identifications were confirmed using Whole Genome Sequencing as below: The sequencing method carried out in this experiment follows the "PCR tiling of SARS-CoV-2 virus with rapid barcoding and Midnight RT PCR Expansion" provided by Oxford Nanopore Technology (Oxford, UK) developed based on a protocol by ARTIC network group 1 . Library preparation process started with reverse transcription, which consists of mixing the purified viral RNA with LunaScript RT SuperMix and incubating the mixtures in a thermal cycler. DNA fragments to be used in the assembly process were amplified by PCR using Midnight primer set (V3) and attached with barcodes from Rapid Barcode Plate (RB96). The mixtures from each sample were pooled together, cleaned with AMPure XP Beads (AXP) and attached with Rapid Adapter F (RAP F). The prepared DNA fragments were then loaded into a primed flow cell (FLO-MIN106) and sequenced on GridION MK1 system.
Viral genome assembly and classification The output sequencing data (.fast5) from MinKNOW software was base-called with Guppy software using the High Accuracy (HAC) model to generate nucleotide sequence data for each fragment (reads) in the fastq format. These base-called data were then processed through the established workflow wf-artic on EPI2ME software to be assembled into consensus sequences. Only reads with average Phred Quality (Q) score above 9 and Adverse events (AE) and Serious Adverse Events See supplementary files for: Supplementary file..
References
Bernal, Da Silva, Musungaie, Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients, N Engl J Med
Bruel, Hadjadj, Maes, Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies, Nat Med
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res
Gottlieb, Vaca, Paredes, Early Remdesivir to Prevent Progression to Severe Covid-19 in Outpatients, N Engl J Med
Hammond, Leister-Tebbe, Gardner, Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19, N Engl J Med
Jittamala, Schilling, Watson, Clinical antiviral efficacy of remdesivir and casirivimab/imdevimab against the SARS-CoV-2 Delta and Omicron variants
Kobylinski, Jittamala, Hanboonkunupakarn, Safety, Pharmacokinetics, and Mosquito-Lethal Effects of Ivermectin in Combination With Dihydroartemisinin-Piperaquine and Primaquine in Healthy Adult Thai Subjects, Clin Pharmacol Ther
Kobylinski, Jittamala, Hanboonkunupakarn, Safety, Pharmacokinetics, and Mosquito-Lethal Effects of Ivermectin in Combination With Dihydroartemisinin-Piperaquine and Primaquine in Healthy Adult Thai Subjects, Clin Pharmacol Ther
Lange, Little, Taylor, Robust Statistical Modeling Using the t Distribution
Lawrence, Meyerowitz-Katz, Heathers, Brown, Sheldrick, The lesson of ivermectin: meta-analyses based on summary data alone are inherently unreliable, Nat Med
Mclean, Rashan, Tran, The fragmented COVID-19 therapeutics research landscape: a living systematic review of clinical trial registrations evaluating priority pharmacological interventions
Mega, Latin America's embrace of an unproven COVID treatment is hindering drug trials, Nature
Mellors, Muñoz, Giorgi, Margolick, Tassoni et al., Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection, Ann Intern Med, doi:10.7326/0003-4819-126-12-199706150-00003
Munoz-Fontela, Widerspick, Albrecht, Advances and gaps in SARS-CoV-2 infection models, PLoS Pathog
Natori, Alghamdi, Tazari, Use of Viral Load as a Surrogate Marker in Clinical Studies of Cytomegalovirus in Solid Organ Transplantation: A Systematic Review and Meta-analysis, Clin Infect Dis, doi:10.1093/cid/cix793
Navarro, Camprubí, Requena-Méndez, Safety of high-dose ivermectin: a systematic review and meta-analysis, J Antimicrob Chemother
O'brien, Forleo-Neto, Musser, Subcutaneous REGEN-COV Antibody Combination to Prevent Covid-19, N Engl J Med
Reis, Silva, Silva, Effect of Early Treatment with Ivermectin among Patients with Covid-19, N Engl J Med
Rnase, Adjustment, adjustment for site & variant; WIP
Road, Bangkok, Tambon Bang Phli Yai, Amphoe Bangplee
Robinson, Liew, Tanner, COVID-19 therapeutics: Challenges and directions for the future, Proc Natl Acad Sci U S A
Sigal, Milo, Jassat, Estimating disease severity of Omicron and Delta SARS-CoV-2 infections, Nat Rev Immunol
Smit, Ochomo, Aljayyoussi, Safety and mosquitocidal efficacy of highdose ivermectin when co-administered with dihydroartemisinin-piperaquine in Kenyan adults with uncomplicated malaria (IVERMAL): a randomised, double-blind, placebocontrolled trial, Lancet Infect Dis
Tipthara, Kobylinski, Godejohann, Hanboonkunupakarn, Roth et al., Identification of the metabolites of ivermectin in humans, Pharmacol Res Perspect
Watson, Kissler, Day, Grad, White, Characterizing SARS-CoV-2 Viral Clearance Kinetics to Improve the Design of Antiviral Pharmacometric Studies, Antimicrob Agents Chemother
Weinreich, Sivapalasingam, Norton, REGEN-COV Antibody Combination and Outcomes in Outpatients with Covid-19, N Engl J Med
Yu, Liao, Yuan, Effectiveness of oseltamivir on disease progression and viral RNA shedding in patients with mild pandemic 2009 influenza A H1N1: opportunistic retrospective study of medical charts in China, BMJ
DOI record:
{
"DOI": "10.7554/elife.83201",
"ISSN": [
"2050-084X"
],
"URL": "http://dx.doi.org/10.7554/eLife.83201",
"abstract": "<jats:p><jats:bold>Background:</jats:bold> There is no generally accepted methodology for <jats:italic>in vivo</jats:italic> assessment of antiviral activity in SARS-CoV-2 infections. Ivermectin has been recommended widely as a treatment of COVID-19, but whether it has clinically significant antiviral activity <jats:italic>in vivo</jats:italic> is uncertain.</jats:p><jats:p><jats:bold>Methods:</jats:bold> In a multicentre open label, randomized, controlled adaptive platform trial, adult patients with early symptomatic COVID-19 were randomized to one of six treatment arms including high dose oral ivermectin (600µg/kg daily for seven days), the monoclonal antibodies casirivimab and imdevimab (600mg/600mg), and no study drug. The primary outcome was the comparison of viral clearance rates in the modified intention-to-treat population (mITT). This was derived from daily log<jats:sub>10</jats:sub> viral densities in standardized duplicate oropharyngeal swab eluates. This ongoing trial is registered at ClinicalTrials.gov (NCT05041907).</jats:p><jats:p><jats:bold>Results:</jats:bold> Randomization to the ivermectin arm was stopped after enrolling 205 patients into all arms, as the prespecified futility threshold was reached. Following ivermectin the mean estimated rate of SARS-CoV-2 viral clearance was 9.1% slower [95%CI -27.2% to +11.8%; n=45] than in the no drug arm [n=41], whereas in a preliminary analysis of the casirivimab/imdevimab arm it was 52.3% faster [95%CI +7.0% to +115.1%; n=10 (Delta variant) versus n=41].</jats:p><jats:p><jats:bold>Conclusions:</jats:bold> High dose ivermectin did not have measurable antiviral activity in early symptomatic COVID-19. Pharmacometric evaluation of viral clearance rate from frequent serial oropharyngeal qPCR viral density estimates is a highly efficient and well tolerated method of assessing SARS CoV-2 antiviral therapeutics <jats:italic>in vivo</jats:italic>.</jats:p><jats:p><jats:bold>Funding:</jats:bold> 'Finding treatments for COVID-19: A phase 2 multi-centre adaptive platform trial to assess antiviral pharmacodynamics in early symptomatic COVID-19 (PLAT-COV)' is supported by the Wellcome Trust Grant ref: 223195/Z/21/Z through the COVID-19 Therapeutics Accelerator.</jats:p><jats:p><jats:bold>Clinical trial number:</jats:bold> ClinicalTrials.gov (NCT05041907).</jats:p>",
"accepted": {
"date-parts": [
[
2023,
2,
3
]
]
},
"alternative-id": [
"10.7554/eLife.83201"
],
"assertion": [
{
"group": {
"name": "peer_review_taxonomy"
},
"label": "Peer review transparency",
"name": "peer_review_transparency",
"value": "single anonymised"
},
{
"group": {
"name": "peer_review_taxonomy"
},
"label": "Peer review interaction",
"name": "peer_review_interaction",
"value": "other reviewer(s), editor"
},
{
"group": {
"name": "peer_review_taxonomy"
},
"label": "Peer review published",
"name": "peer_review_published",
"value": "review summaries, review reports, author/editor communication, reviewer identities reviewer opt in, editor identities"
},
{
"group": {
"name": "post_publication_commenting"
},
"label": "Post publication commenting",
"name": "post_publication_commenting",
"value": "open (sign in with ORCID iD required)"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-6328-8748",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Schilling",
"given": "William HK",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Jittamala",
"given": "Podjanee",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5524-0325",
"affiliation": [
{
"name": "Nuffield Department of Medicine, Oxford University Clinical Research Unit",
"place": [
"Oxford, United Kingdom"
]
}
],
"authenticated-orcid": true,
"family": "Watson",
"given": "James A",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Ekkapongpisit",
"given": "Maneerat",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Tropical Medicine, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Siripoon",
"given": "Tanaya",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Tropical Medicine, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Ngamprasertchai",
"given": "Thundon",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9270-3720",
"affiliation": [
{
"name": "Department of Clinical Tropical Medicine, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Luvira",
"given": "Viravarn",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Pongwilai",
"given": "Sasithorn",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8393-8536",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Cruz",
"given": "Cintia Valeria",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3218-2166",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Callery",
"given": "James J",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Boyd",
"given": "Simon",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Kruabkontho",
"given": "Varaporn",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Ngernseng",
"given": "Thatsanun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Tubprasert",
"given": "Jaruwan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Abdad",
"given": "Mohammad Yazid",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Piaraksa",
"given": "Nattaporn",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Suwannasin",
"given": "Kanokon",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Bangplee Hospital, Ministry of Public Health",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Hanboonkunupakarn",
"given": "Pongtorn",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Hanboonkunupakarn",
"given": "Borimas",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Bangplee Hospital, Ministry of Public Health",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Sookprome",
"given": "Sakol",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Poovorawan",
"given": "Kittiyod",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6184-3381",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Thaipadungpanit",
"given": "Janjira",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Blacksell",
"given": "Stuart",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Imwong",
"given": "Mallika",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4566-4030",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Tarning",
"given": "Joel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Taylor",
"given": "Walter RJ",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Faculty of Medicine, Navamindradhiraj University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Chotivanich",
"given": "Vasin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Faculty of Science and Health Technology, Navamindradhiraj University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Sangketchon",
"given": "Chunlanee",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Faculty of Medicine, Navamindradhiraj University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Ruksakul",
"given": "Wiroj",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Chotivanich",
"given": "Kesinee",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6944-3008",
"affiliation": [
{
"name": "Department of Biochemistry and Immunology, Universidade Federal de Minas Gerais",
"place": [
"Belo Horizonte, Brazil"
]
}
],
"authenticated-orcid": true,
"family": "Teixeira",
"given": "Mauro Martins",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Pukrittayakamee",
"given": "Sasithon",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5190-2395",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Dondorp",
"given": "Arjen M",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2309-1171",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "Day",
"given": "Nicholas PJ",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Tropical Medicine, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Piyaphanee",
"given": "Watcharapong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Tropical Medicine, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"family": "Phumratanaprapin",
"given": "Weerapong",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1897-1978",
"affiliation": [
{
"name": "Mahidol Oxford Tropical Medicine Research Unit, Mahidol University",
"place": [
"Bangkok, Thailand"
]
}
],
"authenticated-orcid": true,
"family": "White",
"given": "Nicholas J",
"sequence": "additional"
},
{
"affiliation": [],
"name": "on behalf of the PLATCOV Collaborative Group",
"sequence": "additional"
}
],
"container-title": "eLife",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"elifesciences.org"
]
},
"created": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T00:00:54Z",
"timestamp": 1676937654000
},
"deposited": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T00:02:48Z",
"timestamp": 1676937768000
},
"funder": [
{
"DOI": "10.13039/100010269",
"award": [
"223195/Z/21/Z"
],
"doi-asserted-by": "publisher",
"name": "Wellcome Trust"
},
{
"DOI": "10.13039/100010269",
"award": [
"223195/Z/21/Z"
],
"doi-asserted-by": "publisher",
"name": "Wellcome Trust"
},
{
"DOI": "10.13039/100010269",
"award": [
"223195/Z/21/Z"
],
"doi-asserted-by": "publisher",
"name": "Wellcome Trust"
}
],
"indexed": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T05:35:51Z",
"timestamp": 1676957751211
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
2,
21
]
]
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T00:00:00Z",
"timestamp": 1676937600000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "am",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T00:00:00Z",
"timestamp": 1676937600000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
2,
21
]
],
"date-time": "2023-02-21T00:00:00Z",
"timestamp": 1676937600000
}
}
],
"link": [
{
"URL": "https://cdn.elifesciences.org/articles/83201/elife-83201-v1.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://cdn.elifesciences.org/articles/83201/elife-83201-v1.xml",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "4374",
"original-title": [],
"prefix": "10.7554",
"published": {
"date-parts": [
[
2023,
2,
21
]
]
},
"published-online": {
"date-parts": [
[
2023,
2,
21
]
]
},
"publisher": "eLife Sciences Publications, Ltd",
"reference-count": 0,
"references-count": 0,
"relation": {
"is-supplemented-by": [
{
"asserted-by": "subject",
"id": "https://github.com/jwatowatson/PLATCOV-Ivermectin",
"id-type": "uri"
}
]
},
"resource": {
"primary": {
"URL": "https://elifesciences.org/articles/83201"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Immunology and Microbiology",
"General Biochemistry, Genetics and Molecular Biology",
"General Medicine",
"General Neuroscience"
],
"subtitle": [],
"title": "Pharmacometrics of high dose ivermectin in early COVID-19: an open label, randomized, controlled adaptive platform trial (PLATCOV)",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.7554/elife.83201",
"volume": "12"
}